Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion
- PMID: 22292984
- PMCID: PMC3297527
- DOI: 10.1186/1471-2407-12-51
Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion
Abstract
Background: miRNAs, endogenous oligonucleotide RNAs, play an important role in mammary gland carcinogenesis and tumor progression. Detection of their expression and investigation of their functions could lead to discovery of novel biomarkers for breast cancer.
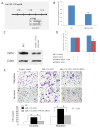
Methods: In situ hybridization was used to detect miR-133a expression in formalin-fixed paraffin-embedded breast surgical specimens from 26 benign, 34 pericancerously normal and 90 cancerous tissues. qRT-PCR was performed to assess miR-133a levels in 6 breast cell lines and 10 benign and 18 cancerous fresh breast tissue specimens. Cell viability, migration, and invasion assays were used to determine the role of miR-133a in regulation of breast cancer cell growth, migration, and invasion, respectively. Luciferase assay was performed to assess miR-133a binding to FSCN1 gene.
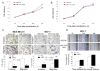
Results: Expression of miR-133a was reduced from normal through benign to cancerous breast tissues. Expression of miR-133a was also low in breast cancer cell lines. The reduced miR-133a expression was associated with lymph nodes metastasis, high clinical stages, and shorter relapse-free survivals of patients with breast cancer. Furthermore, transfection of miR-133a oligonucleotides slightly inhibited growth but significantly decreased migration and invasion capacity of breast cancer cells, compared with negative controls, whereas knockdown of miR-133a expression induced breast cancer cell migration and invasion. In addition, we identified a putative miR-133a binding site in the 3'-untranslated region (UTR) of Fascin1 (FSCN1) gene using an online bioinformatical tool. We found that miR-133a transfection significantly reduced expression of FSCN1 mRNA and protein. The luciferase reporter assay confirmed that FSCN1 was the direct _target gene of miR-133a.
Conclusions: miR-133a expression was lost in breast cancer tissues, loss of which was associated with lymph nodes metastasis, high clinical stages and shorter relapse-free survivals of patients with breast cancer. Functionally, miR-133a can suppress tumor cell invasion and migration and _targeted the expression of FSCN1. Future study will verify whether detection of miR-133a expression can served as a novel biomarker for breast cancer progression and patient prognosis.
Figures

Similar articles
-
miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs _target FSCN1 in esophageal squamous cell carcinoma.Int J Cancer. 2010 Dec 15;127(12):2804-14. doi: 10.1002/ijc.25284. Int J Cancer. 2010. PMID: 21351259
-
MicroRNA-145 and MicroRNA-133a Inhibited Proliferation, Migration, and Invasion, While Promoted Apoptosis in Hepatocellular Carcinoma Cells Via _targeting FSCN1.Dig Dis Sci. 2015 Oct;60(10):3044-52. doi: 10.1007/s10620-015-3706-9. Epub 2015 Jul 15. Dig Dis Sci. 2015. PMID: 26173501
-
MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis.BMC Cancer. 2010 Oct 9;10:542. doi: 10.1186/1471-2407-10-542. BMC Cancer. 2010. PMID: 20932331 Free PMC article.
-
The role and regulatory mechanism of FSCN1 in breast tumorigenesis and progression.Yi Chuan. 2023 Feb 20;45(2):115-127. doi: 10.16288/j.yczz.22-346. Yi Chuan. 2023. PMID: 36927659 Review.
-
Fascin1 in carcinomas: Its regulation and prognostic value.Int J Cancer. 2015 Dec 1;137(11):2534-44. doi: 10.1002/ijc.29260. Epub 2014 Oct 28. Int J Cancer. 2015. PMID: 25302416 Review.
Cited by
-
Altered microRNA expression patterns during the initiation and promotion stages of neonatal diethylstilbestrol-induced dysplasia/neoplasia in the hamster (Mesocricetus auratus) uterus.Cell Biol Toxicol. 2017 Oct;33(5):483-500. doi: 10.1007/s10565-017-9389-6. Epub 2017 Mar 6. Cell Biol Toxicol. 2017. PMID: 28265775 Free PMC article.
-
Transcriptome profiling reveals miR-9-3p as a novel tumor suppressor in gastric cancer.Onco_target. 2017 Jun 6;8(23):37321-37331. doi: 10.18632/onco_target.16310. Onco_target. 2017. PMID: 28418879 Free PMC article.
-
Decreased expression of miR-378 correlates with tumor invasiveness and poor prognosis of patients with glioma.Int J Clin Exp Pathol. 2015 Jun 1;8(6):7016-21. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26261592 Free PMC article.
-
Aberrant expression of a five-microRNA signature in breast carcinoma as a promising biomarker for diagnosis.J Clin Lab Anal. 2020 Feb;34(2):e23063. doi: 10.1002/jcla.23063. Epub 2019 Oct 8. J Clin Lab Anal. 2020. PMID: 31595567 Free PMC article.
-
MicroRNA-133a acts as a tumour suppressor in breast cancer through _targeting LASP1.Oncol Rep. 2018 Feb;39(2):473-482. doi: 10.3892/or.2017.6114. Epub 2017 Nov 27. Oncol Rep. 2018. Retraction in: Oncol Rep. 2022 Jul;48(1):119. doi: 10.3892/or.2022.8330 PMID: 29207145 Free PMC article. Retracted.
References
-
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that _targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. - PubMed
-
- Kawasaki H, Taira K. Retraction: Hes1 is a _target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2 cells. Nature. 2003;426:100. - PubMed
-
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

