Rapid identification of monospecific monoclonal antibodies using a human proteome microarray
- PMID: 22307071
- PMCID: PMC3433917
- DOI: 10.1074/mcp.O111.016253
Rapid identification of monospecific monoclonal antibodies using a human proteome microarray
Abstract
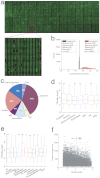
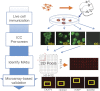
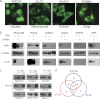
To broaden the range of tools available for proteomic research, we generated a library of 16,368 unique full-length human ORFs that are expressible as N-terminal GST-His(6) fusion proteins. Following expression in yeast, these proteins were then individually purified and used to construct a human proteome microarray. To demonstrate the usefulness of this reagent, we developed a streamlined strategy for the production of monospecific monoclonal antibodies that used immunization with live human cells and microarray-based analysis of antibody specificity as its central components. We showed that microarray-based analysis of antibody specificity can be performed efficiently using a two-dimensional pooling strategy. We also demonstrated that our immunization and selection strategies result in a large fraction of monospecific monoclonal antibodies that are both immunoblot and immunoprecipitation grade. Our data indicate that the pipeline provides a robust platform for the generation of monoclonal antibodies of exceptional specificity.
Figures

Similar articles
-
Preparation and characterization of monoclonal antibody against glypican-3.Hybridoma (Larchmt). 2012 Dec;31(6):455-61. doi: 10.1089/hyb.2012.0030. Hybridoma (Larchmt). 2012. PMID: 23244326
-
High throughput production of mouse monoclonal antibodies using antigen microarrays.Proteomics. 2005 Nov;5(16):4070-81. doi: 10.1002/pmic.200401279. Proteomics. 2005. PMID: 16254927
-
A monoclonal antibody against the catalytic domain of PTP1B.Hybridoma (Larchmt). 2012 Jun;31(3):209-13. doi: 10.1089/hyb.2011.0115. Hybridoma (Larchmt). 2012. PMID: 22741586 Free PMC article.
-
[Production of monoclonal antibodies and use of monospecific and monoclonal antibodies in immunochemical analyses].Fiziol Zh (1978). 1982 Jul-Aug;28(4):477-85. Fiziol Zh (1978). 1982. PMID: 6749561 Review. Russian. No abstract available.
-
A human protein atlas based on antibody proteomics.Curr Opin Mol Ther. 2006 Jun;8(3):185-90. Curr Opin Mol Ther. 2006. PMID: 16774037 Review.
Cited by
-
Affinity proteomics to study endogenous protein complexes: pointers, pitfalls, preferences and perspectives.Biotechniques. 2015 Mar 1;58(3):103-19. doi: 10.2144/000114262. eCollection 2015 Mar. Biotechniques. 2015. PMID: 25757543 Free PMC article.
-
Identification of novel serological autoantibodies in Chinese prostate cancer patients using high-throughput protein arrays.Cancer Immunol Immunother. 2023 Jan;72(1):235-247. doi: 10.1007/s00262-022-03242-0. Epub 2022 Jul 13. Cancer Immunol Immunother. 2023. PMID: 35831618 Free PMC article.
-
Current Experimental Methods for Characterizing Protein-Protein Interactions.ChemMedChem. 2016 Apr 19;11(8):738-56. doi: 10.1002/cmdc.201500495. Epub 2016 Feb 11. ChemMedChem. 2016. PMID: 26864455 Free PMC article. Review.
-
Systematic Prediction of Scaffold Proteins Reveals New Design Principles in Scaffold-Mediated Signal Transduction.PLoS Comput Biol. 2015 Sep 22;11(9):e1004508. doi: 10.1371/journal.pcbi.1004508. eCollection 2015. PLoS Comput Biol. 2015. PMID: 26393507 Free PMC article.
-
Heterogeneous Ribonucleoprotein K (hnRNP K) Binds miR-122, a Mature Liver-Specific MicroRNA Required for Hepatitis C Virus Replication.Mol Cell Proteomics. 2015 Nov;14(11):2878-86. doi: 10.1074/mcp.M115.050344. Epub 2015 Sep 1. Mol Cell Proteomics. 2015. PMID: 26330540 Free PMC article.
References
-
- Uhlén M., (2008) Affinity as a tool in life science. BioTechniques 44, 649–654 - PubMed
-
- Strebhardt K., Ullrich A. (2008) Paul Ehrlich's magic bullet concept: 100 years of progress. Nat. Rev. Cancer 8, 473–480 - PubMed
-
- Köhler G., Milstein C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 - PubMed
-
- Stoevesandt O., Taussig M. J. (2007) Affinity reagent resources for human proteome detection: Initiatives and perspectives. Proteomics 7, 2738–2750 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

