Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model
- PMID: 22365814
- PMCID: PMC3327133
- DOI: 10.1016/j.cell.2012.01.033
Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model
Abstract
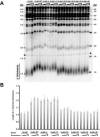
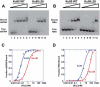
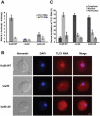
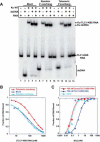
In Saccharomyces cerevisiae, the Ku heterodimer contributes to telomere maintenance as a component of telomeric chromatin and as an accessory subunit of telomerase. How Ku binding to double-stranded DNA (dsDNA) and to telomerase RNA (TLC1) promotes Ku's telomeric functions is incompletely understood. We demonstrate that deletions designed to constrict the DNA-binding ring of Ku80 disrupt nonhomologous end-joining (NHEJ), telomeric gene silencing, and telomere length maintenance, suggesting that these functions require Ku's DNA end-binding activity. Contrary to the current model, a mutant Ku with low affinity for dsDNA also loses affinity for TLC1 both in vitro and in vivo. Competition experiments reveal that wild-type Ku binds dsDNA and TLC1 mutually exclusively. Cells expressing the mutant Ku are deficient in nuclear accumulation of TLC1, as expected from the RNA-binding defect. These findings force reconsideration of the mechanisms by which Ku assists in recruiting telomerase to natural telomeres and broken chromosome ends. PAPERCLIP:
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends.Genes Dev. 2003 Oct 1;17(19):2384-95. doi: 10.1101/gad.1125903. Epub 2003 Sep 15. Genes Dev. 2003. PMID: 12975323 Free PMC article.
-
Loss of Ku's DNA end binding activity affects telomere length via destabilizing telomere-bound Est1 rather than altering TLC1 homeostasis.Sci Rep. 2019 Jul 23;9(1):10607. doi: 10.1038/s41598-019-46840-2. Sci Rep. 2019. PMID: 31337791 Free PMC article.
-
Ku must load directly onto the chromosome end in order to mediate its telomeric functions.PLoS Genet. 2011 Aug;7(8):e1002233. doi: 10.1371/journal.pgen.1002233. Epub 2011 Aug 11. PLoS Genet. 2011. PMID: 21852961 Free PMC article.
-
Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae.Mol Microbiol. 2006 Mar;59(5):1357-68. doi: 10.1111/j.1365-2958.2006.05026.x. Mol Microbiol. 2006. PMID: 16468981 Review.
-
Telomerase: what are the Est proteins doing?Curr Opin Cell Biol. 2003 Jun;15(3):275-80. doi: 10.1016/s0955-0674(03)00040-1. Curr Opin Cell Biol. 2003. PMID: 12787768 Review.
Cited by
-
Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end.Genetics. 2012 Aug;191(4):1073-105. doi: 10.1534/genetics.111.137851. Genetics. 2012. PMID: 22879408 Free PMC article. Review.
-
Ku Binding on Telomeres Occurs at Sites Distal from the Physical Chromosome Ends.PLoS Genet. 2016 Dec 8;12(12):e1006479. doi: 10.1371/journal.pgen.1006479. eCollection 2016 Dec. PLoS Genet. 2016. PMID: 27930670 Free PMC article.
-
Sir4 Deficiency Reverses Cell Senescence by Sub-Telomere Recombination.Cells. 2021 Apr 1;10(4):778. doi: 10.3390/cells10040778. Cells. 2021. PMID: 33915984 Free PMC article.
-
Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo.RNA. 2012 Sep;18(9):1666-78. doi: 10.1261/rna.033555.112. Epub 2012 Jul 31. RNA. 2012. PMID: 22850424 Free PMC article.
-
The Role of Ku70 as a Cytosolic DNA Sensor in Innate Immunity and Beyond.Front Cell Infect Microbiol. 2021 Oct 21;11:761983. doi: 10.3389/fcimb.2021.761983. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34746031 Free PMC article. Review.
References
-
- Antoniacci LM, Kenna MA, Skibbens RV. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6:75–79. - PubMed
-
- Bianchi A, de Lange T. Ku binds telomeric DNA in vitro. J Biol Chem. 1999;274:21223–21227. - PubMed
-
- Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

