miRNA-34c regulates Notch signaling during bone development
- PMID: 22498974
- PMCID: PMC3373245
- DOI: 10.1093/hmg/dds129
miRNA-34c regulates Notch signaling during bone development
Abstract
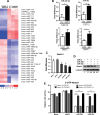
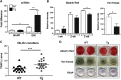
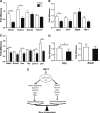
During bone homeostasis, osteoblast and osteoclast differentiation is coupled and regulated by multiple signaling pathways and their downstream transcription factors. Here, we show that microRNA 34 (miR-34) is significantly induced by BMP2 during osteoblast differentiation. In vivo, osteoblast-specific gain of miR-34c in mice leads to an age-dependent osteoporosis due to the defective mineralization and proliferation of osteoblasts and increased osteoclastogenesis. In osteoblasts, miR-34c _targets multiple components of the Notch signaling pathway, including Notch1, Notch2 and Jag1 in a direct manner, and influences osteoclast differentiation in a non-cell-autonomous fashion. Taken together, our results demonstrate that miR-34c is critical during osteoblastogenesis in part by regulating Notch signaling in bone homeostasis. Furthermore, miR-34c-mediated post-transcriptional regulation of Notch signaling in osteoblasts is one possible mechanism to modulate the proliferative effect of Notch in the committed osteoblast progenitors which may be important in the pathogenesis of osteosarcomas. Therefore, understanding the functional interaction of miR-34 and Notch signaling in normal bone development and in bone cancer could potentially lead to therapies modulating miR-34 signaling.
Figures

Similar articles
-
Overexpression of miR-34c inhibits high glucose-induced apoptosis in podocytes by _targeting Notch signaling pathways.Int J Clin Exp Pathol. 2015 May 1;8(5):4525-34. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26191142 Free PMC article.
-
MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells.Stem Cells. 2014 Apr;32(4):902-12. doi: 10.1002/stem.1615. Stem Cells. 2014. PMID: 24307639
-
Maintenance of Bone Homeostasis by DLL1-Mediated Notch Signaling.J Cell Physiol. 2017 Sep;232(9):2569-2580. doi: 10.1002/jcp.25647. Epub 2017 Mar 31. J Cell Physiol. 2017. PMID: 27735989 Free PMC article.
-
Notch and the regulation of osteoclast differentiation and function.Bone. 2020 Sep;138:115474. doi: 10.1016/j.bone.2020.115474. Epub 2020 Jun 8. Bone. 2020. PMID: 32526405 Free PMC article. Review.
-
MicroRNA functions in osteogenesis and dysfunctions in osteoporosis.Curr Osteoporos Rep. 2013 Jun;11(2):72-82. doi: 10.1007/s11914-013-0143-6. Curr Osteoporos Rep. 2013. PMID: 23605904 Free PMC article. Review.
Cited by
-
Epithelial Cell-Derived Extracellular Vesicles Trigger the Differentiation of Two Epithelial Cell Lines.Int J Mol Sci. 2022 Feb 2;23(3):1718. doi: 10.3390/ijms23031718. Int J Mol Sci. 2022. PMID: 35163646 Free PMC article.
-
miRNA-429 suppresses osteogenic differentiation of human adipose-derived mesenchymal stem cells under oxidative stress via _targeting SCD-1.Exp Ther Med. 2020 Jan;19(1):696-702. doi: 10.3892/etm.2019.8246. Epub 2019 Nov 26. Exp Ther Med. 2020. Retraction in: Exp Ther Med. 2022 Jun;23(6):384. doi: 10.3892/etm.2022.11311 PMID: 31885708 Free PMC article. Retracted.
-
LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis.Aging (Albany NY). 2019 Oct 26;11(20):8777-8791. doi: 10.18632/aging.102264. Epub 2019 Oct 26. Aging (Albany NY). 2019. PMID: 31659145 Free PMC article.
-
The microRNAs miR-449a and miR-424 suppress osteosarcoma by _targeting cyclin A2 expression.J Biol Chem. 2019 Mar 22;294(12):4381-4400. doi: 10.1074/jbc.RA118.005778. Epub 2019 Jan 24. J Biol Chem. 2019. PMID: 30679313 Free PMC article.
-
MiR-34b-5p Mediates the Proliferation and Differentiation of Myoblasts by _targeting IGFBP2.Cells. 2019 Apr 17;8(4):360. doi: 10.3390/cells8040360. Cells. 2019. PMID: 30999686 Free PMC article.
References
-
- Gaur T., Lengner C.J., Hovhannisyan H., Bhat R.A., Bodine P.V., Komm B.S., Javed A., van Wijnen A.J., Stein J.L., Stein G.S., et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 2005;280:33132–33140. - PubMed
-
- Lian J.B., Stein G.S., Javed A., van Wijnen A.J., Stein J.L., Montecino M., Hassan M.Q., Gaur T., Lengner C.J., Young D.W. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 2006;7:1–16. - PubMed
-
- Karsenty G. Transcriptional control of skeletogenesis. Annu. Rev. Genomics Hum. Genet. 2008;9:183–196. - PubMed
-
- Deregowski V., Gazzerro E., Priest L., Rydziel S., Canalis E. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J. Biol. Chem. 2006;281:6203–6210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

