HAltORF: a database of predicted out-of-frame alternative open reading frames in human
- PMID: 22613085
- PMCID: PMC3356836
- DOI: 10.1093/database/bas025
HAltORF: a database of predicted out-of-frame alternative open reading frames in human
Abstract
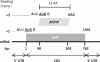
Human alternative open reading frames (HAltORF) is a publicly available and searchable online database referencing putative products of out-of-frame alternative translation initiation (ATI) in human mRNAs. Out-of-frame ATI is a process by which a single mRNA encodes independent proteins, when distinct initiation codons located in different reading frames are recognized by a ribosome to initiate translation. This mechanism is largely used in viruses to increase the coding potential of small viral genomes. There is increasing evidence that out-of-frame ATI is also used in eukaryotes, including human, and may contribute to the diversity of the human proteome. HAltORF is the first web-based searchable database that allows thorough investigation in the human transcriptome of out-of-frame alternative open reading frames with a start codon located in a strong Kozak context, and are thus the more likely to be expressed. It is also the first large scale study on the human transcriptome to successfully predict the expression of out-of-frame ATI protein products that were previously discovered experimentally. HAltORF will be a useful tool for the identification of human genes with multiple coding sequences, and will help to better define and understand the complexity of the human proteome. Database URL: http://haltorf.roucoulab.com/.
Figures

Similar articles
-
Direct detection of alternative open reading frames translation products in human significantly expands the proteome.PLoS One. 2013 Aug 12;8(8):e70698. doi: 10.1371/journal.pone.0070698. eCollection 2013. PLoS One. 2013. PMID: 23950983 Free PMC article.
-
uPEPperoni: an online tool for upstream open reading frame location and analysis of transcript conservation.BMC Bioinformatics. 2014 Feb 1;15:36. doi: 10.1186/1471-2105-15-36. BMC Bioinformatics. 2014. PMID: 24484385 Free PMC article.
-
Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein.Nucleic Acids Res. 2005 Mar 8;33(5):1474-86. doi: 10.1093/nar/gki292. Print 2005. Nucleic Acids Res. 2005. PMID: 15755749 Free PMC article.
-
Alternative translation start sites and hidden coding potential of eukaryotic mRNAs.Bioessays. 2008 Jul;30(7):683-91. doi: 10.1002/bies.20771. Bioessays. 2008. PMID: 18536038 Review.
-
Translational Regulation by Upstream Open Reading Frames and Human Diseases.Adv Exp Med Biol. 2019;1157:99-116. doi: 10.1007/978-3-030-19966-1_5. Adv Exp Med Biol. 2019. PMID: 31342439 Review.
Cited by
-
Proteogenomics: Integrating Next-Generation Sequencing and Mass Spectrometry to Characterize Human Proteomic Variation.Annu Rev Anal Chem (Palo Alto Calif). 2016 Jun 12;9(1):521-45. doi: 10.1146/annurev-anchem-071015-041722. Epub 2016 Mar 30. Annu Rev Anal Chem (Palo Alto Calif). 2016. PMID: 27049631 Free PMC article. Review.
-
Proteogenomics from a bioinformatics angle: A growing field.Mass Spectrom Rev. 2017 Sep;36(5):584-599. doi: 10.1002/mas.21483. Epub 2015 Dec 15. Mass Spectrom Rev. 2017. PMID: 26670565 Free PMC article. Review.
-
OCCAM: prediction of small ORFs in bacterial genomes by means of a _target-decoy database approach and machine learning techniques.Database (Oxford). 2020 Jan 1;2020:baaa067. doi: 10.1093/database/baaa067. Database (Oxford). 2020. PMID: 33206960 Free PMC article.
-
Origin, Evolution and Stability of Overlapping Genes in Viruses: A Systematic Review.Genes (Basel). 2021 May 26;12(6):809. doi: 10.3390/genes12060809. Genes (Basel). 2021. PMID: 34073395 Free PMC article.
-
The influence of transcript assembly on the proteogenomics discovery of microproteins.PLoS One. 2018 Mar 27;13(3):e0194518. doi: 10.1371/journal.pone.0194518. eCollection 2018. PLoS One. 2018. PMID: 29584760 Free PMC article.
References
-
- Kochetov AV. Alternative translation start sites and their significance for eukaryotic proteome. Mol. Biol. (Mosk) 2006;40:788–795. - PubMed
-
- Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. - PubMed
-
- Veloso F, Riadi G, Aliaga D, et al. Large-scale, multi-genome analysis of alternate open reading frames in bacteria and archaea. Omics. 2005;9:91–105. - PubMed
-
- Branch AD, Stump DD, Gutierrez JA, et al. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 2005;25:105–117. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

