Plac1 (placenta-specific 1) is essential for normal placental and embryonic development
- PMID: 22729990
- PMCID: PMC4594876
- DOI: 10.1002/mrd.22062
Plac1 (placenta-specific 1) is essential for normal placental and embryonic development
Abstract
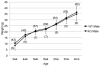
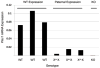
Plac1 is a recently identified, X-linked gene whose expression is restricted primarily to cells of the trophoblast lineage. It localizes to a chromosomal locus previously implicated in placental growth. We therefore sought to determine if Plac1 is necessary for placental and embryonic development by examining a mutant mouse model. Plac1 ablation resulted in placentomegaly and mild intrauterine growth retardation (IUGR). At E16.5, knockout (KO) and heterozygous (Het) placentae of the Plac1-null allele inherited from the mother (X(m-) X) weighed approximately 100% more than wildtype (WT) placentae, whereas the corresponding embryos weighed 7-12% less. Histologically, Plac1 mutants exhibited an expanded spongiotrophoblast layer that invaded the labyrinth. By contrast, Het placentae that inherited the null allele from the father (XX(p-) ) exhibited normal growth and were histologically indistinguishable from WT placentae, consistent with paternal imprinting of Plac1. When examined across gestation, WT and X(m-) X placental weights peaked at E16.5 and decreased slightly thereafter. KO placentae (X(m-) X(p-) and X(m-) Y), however, continued to increase in weight after E16.5, consistent with a functional role for the paternal Plac1 allele. Subsequent analysis confirmed that the paternal allele partially escapes complete X-inactivation and thus contributes to placental growth regulation. Additionally, although male Plac1 KO mice can survive, they exhibit decreased viability as a consequence of events occurring late in gestation or shortly after birth. Thus, Plac1 is a paternally imprinted, X-linked gene essential for normal placental and embryonic development.
Copyright © 2012 Wiley Periodicals, Inc.
Figures




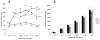

Similar articles
-
Plac1 (placenta-specific 1) is widely expressed during fetal development and is associated with a lethal form of hydrocephalus.Birth Defects Res A Clin Mol Teratol. 2013 Sep;97(9):571-7. doi: 10.1002/bdra.23171. Epub 2013 Sep 6. Birth Defects Res A Clin Mol Teratol. 2013. PMID: 24014101
-
Lentiviral Vector-Mediated Complementation Restored Fetal Viability but Not Placental Hyperplasia in Plac1-Deficient Mice.Biol Reprod. 2016 Jan;94(1):6. doi: 10.1095/biolreprod.115.133454. Epub 2015 Nov 19. Biol Reprod. 2016. PMID: 26586843
-
PLAC1, a trophoblast-specific gene, is expressed throughout pregnancy in the human placenta and modulated by keratinocyte growth factor.Mol Reprod Dev. 2002 Dec;63(4):430-6. doi: 10.1002/mrd.10200. Mol Reprod Dev. 2002. PMID: 12412044
-
PLAC1 (Placenta-specific 1): a novel, X-linked gene with roles in reproductive and cancer biology.Prenat Diagn. 2010 Jun;30(6):497-502. doi: 10.1002/pd.2506. Prenat Diagn. 2010. PMID: 20509147 Free PMC article. Review.
-
[Meiotic inactivation of sex chromosomes in mammals].Genetika. 2010 Apr;46(4):437-47. Genetika. 2010. PMID: 20536013 Review. Russian.
Cited by
-
Identification of placenta-specific protein 1 (PLAC-1) expression on human PC-3 cell line-derived prostate cancer stem cells compared to the tumor parental cells.Discov Oncol. 2024 Jun 28;15(1):251. doi: 10.1007/s12672-024-01121-x. Discov Oncol. 2024. PMID: 38943028 Free PMC article.
-
Fetal growth delay caused by loss of non-canonical imprinting is resolved late in pregnancy and culminates in offspring overgrowth.Elife. 2024 May 30;13:e81875. doi: 10.7554/eLife.81875. Elife. 2024. PMID: 38813868 Free PMC article.
-
Placenta: an old organ with new functions.Front Immunol. 2024 Apr 19;15:1385762. doi: 10.3389/fimmu.2024.1385762. eCollection 2024. Front Immunol. 2024. PMID: 38707901 Free PMC article. Review.
-
Placental adaptations supporting fetal growth during normal and adverse gestational environments.Exp Physiol. 2023 Mar;108(3):371-397. doi: 10.1113/EP090442. Epub 2022 Dec 9. Exp Physiol. 2023. PMID: 36484327 Free PMC article. Review.
-
The X-linked splicing regulator MBNL3 has been co-opted to restrict placental growth in eutherians.PLoS Biol. 2022 Apr 27;20(4):e3001615. doi: 10.1371/journal.pbio.3001615. eCollection 2022 Apr. PLoS Biol. 2022. PMID: 35476669 Free PMC article.
References
-
- Cocchia M, Huber R, Pantano S, Chen EY, Ma P, Forabosco A, Ko MS, Schlessinger D. PLAC1, an Xq26 gene with placenta-specific expression. Genomics. 2000;68:305–12. - PubMed
-
- Fant M, Weisoly DL, Cocchia M, Huber R, Khan S, Lunt T, Schlessinger D. PLAC1, a trophoblast-specific gene, is expressed throughout pregnancy in the human placenta and modulated by keratinocyte growth factor. Mol Reprod Dev. 2002;63:430–6. - PubMed
-
- Fant M, Barerra-Saldana H, Dubinsky W, Poindexter B, Bick R. The PLAC1 protein localizes to membranous compartments in the apical region of the syncytiotrophoblast. Mol Reprod Dev. 2007;74:922–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

