Compromised fertility disrupts Peg1 but not Snrpn and Peg3 imprinted methylation acquisition in mouse oocytes
- PMID: 22798963
- PMCID: PMC3394371
- DOI: 10.3389/fgene.2012.00129
Compromised fertility disrupts Peg1 but not Snrpn and Peg3 imprinted methylation acquisition in mouse oocytes
Abstract
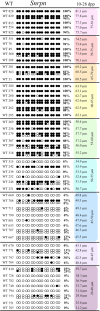
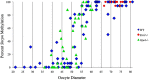
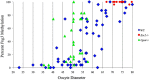
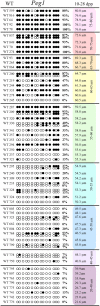
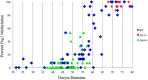
Growth and maturation of healthy oocytes within follicles requires bidirectional signaling and intercellular gap junctional communication. Aberrant endocrine signaling and loss of gap junctional communication between the oocyte and granulosa cells leads to compromised folliculogenesis, oocyte maturation, and oocyte competency, consequently impairing fertility. Given that oocyte-specific DNA methylation establishment at imprinted genes occurs during this growth phase, we determined whether compromised endocrine signaling and gap junctional communication would disrupt de novo methylation acquisition using ERβ and connexin37 genetic models. To compare mutant oocytes to control oocytes, DNA methylation acquisition was first examined in individual, 20-80 μm control oocytes at three imprinted genes, Snrpn, Peg3, and Peg1. We observed that each gene has its own size-dependent acquisition kinetics, similar to previous studies. To determine whether compromised endocrine signaling and gap junctional communication disrupted de novo methylation acquisition,individual oocytes from Esr2- and Gja4-deficient mice were also assessed for DNA methylation establishment. We observed no aberrant or delayed acquisition of DNA methylation at Snrpn, Peg3, or Peg1 in oocytes from Esr2-deficient females, and no perturbation in Snrpn or Peg3de novo methylation in oocytes from Gja4-null females. However, Gja4 deficiency resulted in a loss or delay in methylation acquisition at Peg1. One explanation for this difference between the three loci analyzed is the late establishment of DNA methylation at the Peg1 gene. These results indicate that compromised fertility though impaired intercellular communication can lead to imprinting acquisition errors. Further studies are required to determine the effects of subfertility/infertility originating from impaired signaling and intercellular communication during oogenesis on imprint maintenance during preimplantation development.
Keywords: DNA methylation; connexin37; estrogen receptor beta; genomic imprinting; imprint acquisition; infertility; oocyte; oocyte diameter.
Figures





Similar articles
-
Human in vitro oocyte maturation is not associated with increased imprinting error rates at LIT1, SNRPN, PEG3 and GTL2.Hum Reprod. 2014 Sep;29(9):1995-2005. doi: 10.1093/humrep/deu155. Epub 2014 Jun 24. Hum Reprod. 2014. PMID: 24963167
-
Methylation dynamics of imprinted genes in mouse germ cells.Genomics. 2002 Apr;79(4):530-8. doi: 10.1006/geno.2002.6732. Genomics. 2002. PMID: 11944985
-
Effect of postovulatory oocyte aging on DNA methylation imprinting acquisition in offspring oocytes.Fertil Steril. 2011 Dec;96(6):1479-84. doi: 10.1016/j.fertnstert.2011.09.022. Epub 2011 Oct 6. Fertil Steril. 2011. PMID: 21982284
-
Maintenance of Mest imprinted methylation in blastocyst-stage mouse embryos is less stable than other imprinted loci following superovulation or embryo culture.Environ Epigenet. 2017 Aug 29;3(3):dvx015. doi: 10.1093/eep/dvx015. eCollection 2017 Jul. Environ Epigenet. 2017. PMID: 29492315 Free PMC article. Review.
-
Culture of oocytes and risk of imprinting defects.Hum Reprod Update. 2013 Jan-Feb;19(1):52-66. doi: 10.1093/humupd/dms042. Epub 2012 Oct 10. Hum Reprod Update. 2013. PMID: 23054129 Review.
Cited by
-
Maternal effect factors that contribute to oocytes developmental competence: an update.J Assist Reprod Genet. 2022 Apr;39(4):861-871. doi: 10.1007/s10815-022-02434-y. Epub 2022 Feb 15. J Assist Reprod Genet. 2022. PMID: 35165782 Free PMC article. Review.
-
Epigenetic changes in mammalian gametes throughout their lifetime: the four seasons metaphor.Chromosoma. 2019 Sep;128(3):423-441. doi: 10.1007/s00412-019-00704-w. Epub 2019 Apr 27. Chromosoma. 2019. PMID: 31030260 Review.
-
Transcription and chromatin determinants of de novo DNA methylation timing in oocytes.Epigenetics Chromatin. 2017 May 12;10:25. doi: 10.1186/s13072-017-0133-5. eCollection 2017. Epigenetics Chromatin. 2017. PMID: 28507606 Free PMC article.
-
An overview of gene expression dynamics during early ovarian folliculogenesis: specificity of follicular compartments and bi-directional dialog.BMC Genomics. 2013 Dec 19;14:904. doi: 10.1186/1471-2164-14-904. BMC Genomics. 2013. PMID: 24350644 Free PMC article.
-
Bisphenol A Effects on Mammalian Oogenesis and Epigenetic Integrity of Oocytes: A Case Study Exploring Risks of Endocrine Disrupting Chemicals.Biomed Res Int. 2015;2015:698795. doi: 10.1155/2015/698795. Epub 2015 Aug 3. Biomed Res Int. 2015. PMID: 26339634 Free PMC article. Review.
References
-
- Anckaert E., Adriaenssens T., Romero S., Dremier S., Smitz J. (2009). Unaltered imprinting establishment of key imprinted genes in mouse oocytes after in vitro follicle culture under variable follicle-stimulating hormone exposure. Int. J. Dev. Biol. 53 541–548 - PubMed
-
- Anckaert E., Romero S., Adriaenssens T., Smitz J. (2010). Effects of low methyl donor levels in culture medium during mouse follicle culture on oocyte imprinting establishment. Biol. Reprod. 83 377–386 - PubMed
-
- Azzi S., Rossignol S., Steunou V., Sas T., Thibaud N., Danton F., Le Jule M., Heinrichs C., Cabrol S., Gicquel C., Le Bouc Y., Netchine I. (2009). Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell–Silver and Beckwith–Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum. Mol. Genet. 18 4724–4733 - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

