Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes
- PMID: 22873405
- PMCID: PMC3539919
- DOI: 10.1186/1475-2859-11-103
Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes
Abstract
Background: Pichia pastoris is an established eukaryotic host for the production of recombinant proteins. Most often, protein production is under the control of the strong methanol-inducible aox1 promoter. However, detailed information about the physiological alterations in P. pastoris accompanying the shift from growth on glycerol to methanol-induced protein production under industrial relevant conditions is missing. Here, we provide an analysis of the physiological response of P. pastoris GS115 to methanol-induced high-level production of the Hepatitis B virus surface antigen (HBsAg). High product titers and the retention of the protein in the endoplasmic reticulum (ER) are supposedly of major impact on the host physiology. For a more detailed understanding of the cellular response to methanol-induced HBsAg production, the time-dependent changes in the yeast proteome and ultrastructural cell morphology were analyzed during the production process.
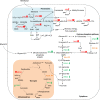
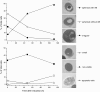
Results: The shift from growth on glycerol to growth and HBsAg production on methanol was accompanied by a drastic change in the yeast proteome. In particular, enzymes from the methanol dissimilation pathway started to dominate the proteome while enzymes from the methanol assimilation pathway, e.g. the transketolase DAS1, increased only moderately. The majority of methanol was metabolized via the energy generating dissimilatory pathway leading to a corresponding increase in mitochondrial size and numbers. The methanol-metabolism related generation of reactive oxygen species induced a pronounced oxidative stress response (e.g. strong increase of the peroxiredoxin PMP20). Moreover, the accumulation of HBsAg in the ER resulted in the induction of the unfolded protein response (e.g. strong increase of the ER-resident disulfide isomerase, PDI) and the ER associated degradation (ERAD) pathway (e.g. increase of two cytosolic chaperones and members of the AAA ATPase superfamily) indicating that potential degradation of HBsAg could proceed via the ERAD pathway and through the proteasome. However, the amount of HBsAg did not show any significant decline during the cultivation revealing its general protection from proteolytic degradation. During the methanol fed-batch phase, induction of vacuolar proteases (e.g. strong increase of APR1) and constitutive autophagic processes were observed. Vacuolar enclosures were mainly found around peroxisomes and not close to HBsAg deposits and, thus, were most likely provoked by peroxisomal components damaged by reactive oxygen species generated by methanol oxidation.
Conclusions: In the methanol fed-batch phase P. pastoris is exposed to dual stress; stress resulting from methanol degradation and stress resulting from the production of the recombinant protein leading to the induction of oxidative stress and unfolded protein response pathways, respectively. Finally, the modest increase of methanol assimilatory enzymes compared to the strong increase of methanol dissimilatory enzymes suggests here a potential to increase methanol incorporation into biomass/product through metabolic enhancement of the methanol assimilatory pathway.
Figures



Similar articles
-
Decrease of UPR- and ERAD-related proteins in Pichia pastoris during methanol-induced secretory insulin precursor production in controlled fed-batch cultures.Microb Cell Fact. 2014 Feb 13;13(1):23. doi: 10.1186/1475-2859-13-23. Microb Cell Fact. 2014. PMID: 24521445 Free PMC article.
-
Fate of the UPR marker protein Kar2/Bip and autophagic processes in fed-batch cultures of secretory insulin precursor producing Pichia pastoris.Microb Cell Fact. 2018 Aug 9;17(1):123. doi: 10.1186/s12934-018-0970-3. Microb Cell Fact. 2018. PMID: 30092809 Free PMC article.
-
Expression of hepatitis B surface antigen in the methylotrophic yeast Pichia pastoris using the GAP promoter.J Biotechnol. 2001 Jun 1;88(1):21-35. doi: 10.1016/s0168-1656(01)00254-1. J Biotechnol. 2001. PMID: 11377762
-
Engineering of the unfolded protein response pathway in Pichia pastoris: enhancing production of secreted recombinant proteins.Appl Microbiol Biotechnol. 2021 Jun;105(11):4397-4414. doi: 10.1007/s00253-021-11336-5. Epub 2021 May 26. Appl Microbiol Biotechnol. 2021. PMID: 34037840 Free PMC article. Review.
-
Molecular chaperones and proteostasis regulation during redox imbalance.Redox Biol. 2014 Jan 30;2:323-32. doi: 10.1016/j.redox.2014.01.017. eCollection 2014. Redox Biol. 2014. PMID: 24563850 Free PMC article. Review.
Cited by
-
Comparative transcriptome and metabolome analyses reveal the methanol dissimilation pathway of Pichia pastoris.BMC Genomics. 2022 May 12;23(1):366. doi: 10.1186/s12864-022-08592-8. BMC Genomics. 2022. PMID: 35549850 Free PMC article.
-
Yeast Expression Systems: Overview and Recent Advances.Mol Biotechnol. 2019 May;61(5):365-384. doi: 10.1007/s12033-019-00164-8. Mol Biotechnol. 2019. PMID: 30805909 Review.
-
Molecular optimization of rabies virus glycoprotein expression in Pichia pastoris.Microb Biotechnol. 2016 May;9(3):355-68. doi: 10.1111/1751-7915.12350. Epub 2016 Feb 16. Microb Biotechnol. 2016. PMID: 26880068 Free PMC article.
-
Avoiding overflow metabolite formation in Komagataella phaffii fermentations to enhance recombinant protein production.J Biol Eng. 2024 Oct 3;18(1):54. doi: 10.1186/s13036-024-00453-0. J Biol Eng. 2024. PMID: 39363343 Free PMC article.
-
Characterization of the off-flavor from Pichia pastoris GS115 during the overexpression of an α-l-rhamnosidase.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuad035. doi: 10.1093/jimb/kuad035. J Ind Microbiol Biotechnol. 2023. PMID: 37942557 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

