Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase
- PMID: 22902005
- PMCID: PMC3491369
- DOI: 10.1186/gb-2012-13-8-r69
Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase
Abstract
Background: Methylation of cytosine in DNA (5mC) is an important epigenetic mark that is involved in the regulation of genome function. During early embryonic development in mammals, the methylation landscape is dynamically reprogrammed in part through active demethylation. Recent advances have identified key players involved in active demethylation pathways, including oxidation of 5mC to 5-hydroxymethylcytosine (5hmC) and 5-formylcytosine (5fC) by the TET enzymes, and excision of 5fC by the base excision repair enzyme thymine DNA glycosylase (TDG). Here, we provide the first genome-wide map of 5fC in mouse embryonic stem (ES) cells and evaluate potential roles for 5fC in differentiation.
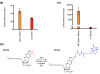
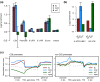
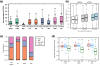
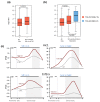
Results: Our method exploits the unique reactivity of 5fC for pulldown and high-throughput sequencing. Genome-wide mapping revealed 5fC enrichment in CpG islands (CGIs) of promoters and exons. CGI promoters in which 5fC was relatively more enriched than 5mC or 5hmC corresponded to transcriptionally active genes. Accordingly, 5fC-rich promoters had elevated H3K4me3 levels, associated with active transcription, and were frequently bound by RNA polymerase II. TDG down-regulation led to 5fC accumulation in CGIs in ES cells, which correlates with increased methylation in these genomic regions during differentiation of ES cells in wild-type and TDG knockout contexts.
Conclusions: Collectively, our data suggest that 5fC plays a role in epigenetic reprogramming within specific genomic regions, which is controlled in part by TDG-mediated excision. Notably, 5fC excision in ES cells is necessary for the correct establishment of CGI methylation patterns during differentiation and hence for appropriate patterns of gene expression during development.
Figures

Similar articles
-
Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation.Nature. 2011 May 19;473(7347):398-402. doi: 10.1038/nature10008. Epub 2011 Apr 3. Nature. 2011. PMID: 21460836
-
Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics.Cell. 2013 Apr 25;153(3):692-706. doi: 10.1016/j.cell.2013.04.002. Epub 2013 Apr 18. Cell. 2013. PMID: 23602152 Free PMC article.
-
In vivo genome-wide profiling reveals a tissue-specific role for 5-formylcytosine.Genome Biol. 2016 Jun 29;17(1):141. doi: 10.1186/s13059-016-1001-5. Genome Biol. 2016. PMID: 27356509 Free PMC article.
-
Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review.
-
MicroRNAs mediated _targeting on the Yin-yang dynamics of DNA methylation in disease and development.Int J Biochem Cell Biol. 2015 Oct;67:115-20. doi: 10.1016/j.biocel.2015.05.002. Epub 2015 May 12. Int J Biochem Cell Biol. 2015. PMID: 25979370 Review.
Cited by
-
Epigenetic Changes in Diabetes and Cardiovascular Risk.Circ Res. 2016 May 27;118(11):1706-22. doi: 10.1161/CIRCRESAHA.116.306819. Circ Res. 2016. PMID: 27230637 Free PMC article. Review.
-
5-Hydroxymethyl-, 5-Formyl- and 5-Carboxydeoxycytidines as Oxidative Lesions and Epigenetic Marks.Chemistry. 2021 Jun 1;27(31):8100-8104. doi: 10.1002/chem.202100551. Epub 2021 May 1. Chemistry. 2021. PMID: 33769637 Free PMC article.
-
Genome-wide analysis reveals a role for TDG in estrogen receptor-mediated enhancer RNA transcription and 3-dimensional reorganization.Epigenetics Chromatin. 2018 Jan 29;11(1):5. doi: 10.1186/s13072-018-0176-2. Epigenetics Chromatin. 2018. PMID: 29378668 Free PMC article.
-
Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer's disease brain.Neurobiol Aging. 2014 Aug;35(8):1850-4. doi: 10.1016/j.neurobiolaging.2014.02.002. Epub 2014 Feb 6. Neurobiol Aging. 2014. PMID: 24679604 Free PMC article.
-
TET proteins: on the frenetic hunt for new cytosine modifications.Brief Funct Genomics. 2013 May;12(3):191-204. doi: 10.1093/bfgp/elt010. Epub 2013 Apr 26. Brief Funct Genomics. 2013. PMID: 23625996 Free PMC article. Review.
References
-
- Pfaffeneder T, Hackner B, Truß M, Münzel M, Müller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell. Angewandte Chemie. 2011;50:7146–7150. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

