Annotation of functional variation in personal genomes using RegulomeDB
- PMID: 22955989
- PMCID: PMC3431494
- DOI: 10.1101/gr.137323.112
Annotation of functional variation in personal genomes using RegulomeDB
Abstract
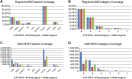
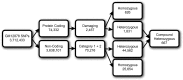
As the sequencing of healthy and disease genomes becomes more commonplace, detailed annotation provides interpretation for individual variation responsible for normal and disease phenotypes. Current approaches focus on direct changes in protein coding genes, particularly nonsynonymous mutations that directly affect the gene product. However, most individual variation occurs outside of genes and, indeed, most markers generated from genome-wide association studies (GWAS) identify variants outside of coding segments. Identification of potential regulatory changes that perturb these sites will lead to a better localization of truly functional variants and interpretation of their effects. We have developed a novel approach and database, RegulomeDB, which guides interpretation of regulatory variants in the human genome. RegulomeDB includes high-throughput, experimental data sets from ENCODE and other sources, as well as computational predictions and manual annotations to identify putative regulatory potential and identify functional variants. These data sources are combined into a powerful tool that scores variants to help separate functional variants from a large pool and provides a small set of putative sites with testable hypotheses as to their function. We demonstrate the applicability of this tool to the annotation of noncoding variants from 69 full sequenced genomes as well as that of a personal genome, where thousands of functionally associated variants were identified. Moreover, we demonstrate a GWAS where the database is able to quickly identify the known associated functional variant and provide a hypothesis as to its function. Overall, we expect this approach and resource to be valuable for the annotation of human genome sequences.
Figures


Similar articles
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
Deep sequencing of Danish Holstein dairy cattle for variant detection and insight into potential loss-of-function variants in protein coding genes.BMC Genomics. 2015 Dec 9;16:1043. doi: 10.1186/s12864-015-2249-y. BMC Genomics. 2015. PMID: 26645365 Free PMC article.
-
Functional annotation signatures of disease susceptibility loci improve SNP association analysis.BMC Genomics. 2014 May 24;15(1):398. doi: 10.1186/1471-2164-15-398. BMC Genomics. 2014. PMID: 24886216 Free PMC article.
-
Incorporating Non-Coding Annotations into Rare Variant Analysis.PLoS One. 2016 Apr 29;11(4):e0154181. doi: 10.1371/journal.pone.0154181. eCollection 2016. PLoS One. 2016. PMID: 27128317 Free PMC article.
-
The genetic basis of systemic lupus erythematosus: What are the risk factors and what have we learned.J Autoimmun. 2016 Nov;74:161-175. doi: 10.1016/j.jaut.2016.08.001. Epub 2016 Aug 10. J Autoimmun. 2016. PMID: 27522116 Review.
Cited by
-
Transcriptional Silencers: Driving Gene Expression with the Brakes On.Trends Genet. 2021 Jun;37(6):514-527. doi: 10.1016/j.tig.2021.02.002. Epub 2021 Mar 9. Trends Genet. 2021. PMID: 33712326 Free PMC article. Review.
-
Novel bayes factors that capture expert uncertainty in prior density specification in genetic association studies.Genet Epidemiol. 2015 May;39(4):239-48. doi: 10.1002/gepi.21891. Epub 2015 Feb 27. Genet Epidemiol. 2015. PMID: 25727067 Free PMC article.
-
Potentially functional variants of ERAP1, PSMF1 and NCF2 in the MHC-I-related pathway predict non-small cell lung cancer survival.Cancer Immunol Immunother. 2021 Oct;70(10):2819-2833. doi: 10.1007/s00262-021-02877-9. Epub 2021 Mar 2. Cancer Immunol Immunother. 2021. PMID: 33651148 Free PMC article.
-
Chromosome X-wide association study in case control studies of pathologically confirmed Alzheimer's disease in a European population.Transl Psychiatry. 2024 Sep 4;14(1):358. doi: 10.1038/s41398-024-03058-9. Transl Psychiatry. 2024. PMID: 39231932 Free PMC article.
-
Mer-tyrosine kinase: a novel susceptibility gene for SLE related end-stage renal disease.Lupus Sci Med. 2022 Nov;9(1):e000752. doi: 10.1136/lupus-2022-000752. Lupus Sci Med. 2022. PMID: 36332927 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
