The methylomes of six bacteria
- PMID: 23034806
- PMCID: PMC3526280
- DOI: 10.1093/nar/gks891
The methylomes of six bacteria
Erratum in
- Nucleic Acids Res. 2014 Apr;42(6):4140
Abstract
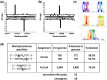
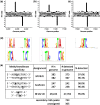
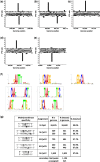
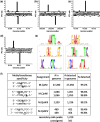
Six bacterial genomes, Geobacter metallireducens GS-15, Chromohalobacter salexigens, Vibrio breoganii 1C-10, Bacillus cereus ATCC 10987, Campylobacter jejuni subsp. jejuni 81-176 and C. jejuni NCTC 11168, all of which had previously been sequenced using other platforms were re-sequenced using single-molecule, real-time (SMRT) sequencing specifically to analyze their methylomes. In every case a number of new N(6)-methyladenine ((m6)A) and N(4)-methylcytosine ((m4)C) methylation patterns were discovered and the DNA methyltransferases (MTases) responsible for those methylation patterns were assigned. In 15 cases, it was possible to match MTase genes with MTase recognition sequences without further sub-cloning. Two Type I restriction systems required sub-cloning to differentiate their recognition sequences, while four MTase genes that were not expressed in the native organism were sub-cloned to test for viability and recognition sequences. Two of these proved active. No attempt was made to detect 5-methylcytosine ((m5)C) recognition motifs from the SMRT® sequencing data because this modification produces weaker signals using current methods. However, all predicted (m6)A and (m4)C MTases were detected unambiguously. This study shows that the addition of SMRT sequencing to traditional sequencing approaches gives a wealth of useful functional information about a genome showing not only which MTase genes are active but also revealing their recognition sequences.
Figures


Similar articles
-
Characterizing the DNA Methyltransferases of Haloferax volcanii via Bioinformatics, Gene Deletion, and SMRT Sequencing.Genes (Basel). 2018 Feb 27;9(3):129. doi: 10.3390/genes9030129. Genes (Basel). 2018. PMID: 29495512 Free PMC article.
-
Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes.Nat Commun. 2015 Jun 15;6:7438. doi: 10.1038/ncomms8438. Nat Commun. 2015. PMID: 26074426 Free PMC article.
-
Methods for Genome-Wide Methylome Profiling of Campylobacter jejuni.Methods Mol Biol. 2017;1512:199-210. doi: 10.1007/978-1-4939-6536-6_17. Methods Mol Biol. 2017. PMID: 27885609
-
The Helicobacter pylori Methylome: Roles in Gene Regulation and Virulence.Curr Top Microbiol Immunol. 2017;400:105-127. doi: 10.1007/978-3-319-50520-6_5. Curr Top Microbiol Immunol. 2017. PMID: 28124151 Review.
-
Diversity of DNA methyltransferases that recognize asymmetric _target sequences.Crit Rev Biochem Mol Biol. 2010 Apr;45(2):125-45. doi: 10.3109/10409231003628007. Crit Rev Biochem Mol Biol. 2010. PMID: 20184512 Review.
Cited by
-
The advantages of SMRT sequencing.Genome Biol. 2013 Jul 3;14(7):405. doi: 10.1186/gb-2013-14-6-405. Genome Biol. 2013. PMID: 23822731 Free PMC article.
-
Actinobacillus pleuropneumoniae encodes multiple phase-variable DNA methyltransferases that control distinct phasevarions.Nucleic Acids Res. 2023 Apr 24;51(7):3240-3260. doi: 10.1093/nar/gkad091. Nucleic Acids Res. 2023. PMID: 36840716 Free PMC article.
-
Enhanced transformation efficiency in Treponema denticola enabled by SyngenicDNA-based plasmids lacking restriction-modification _target motifs.Mol Oral Microbiol. 2023 Dec;38(6):455-470. doi: 10.1111/omi.12441. Epub 2023 Oct 25. Mol Oral Microbiol. 2023. PMID: 37880921 Free PMC article.
-
Super-resolution optical DNA Mapping via DNA methyltransferase-directed click chemistry.Nucleic Acids Res. 2014 Apr;42(7):e50. doi: 10.1093/nar/gkt1406. Epub 2014 Jan 21. Nucleic Acids Res. 2014. PMID: 24452797 Free PMC article.
-
Characterizing the DNA Methyltransferases of Haloferax volcanii via Bioinformatics, Gene Deletion, and SMRT Sequencing.Genes (Basel). 2018 Feb 27;9(3):129. doi: 10.3390/genes9030129. Genes (Basel). 2018. PMID: 29495512 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

