Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells
- PMID: 23045694
- PMCID: PMC3497753
- DOI: 10.1073/pnas.1215899109
Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells
Abstract
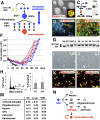
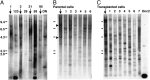
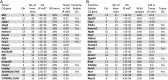
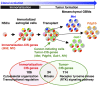
Neural stem cells (NSCs) are considered to be the cell of origin of glioblastoma multiforme (GBM). However, the genetic alterations that transform NSCs into glioma-initiating cells remain elusive. Using a unique transposon mutagenesis strategy that mutagenizes NSCs in culture, followed by additional rounds of mutagenesis to generate tumors in vivo, we have identified genes and signaling pathways that can transform NSCs into glioma-initiating cells. Mobilization of Sleeping Beauty transposons in NSCs induced the immortalization of astroglial-like cells, which were then able to generate tumors with characteristics of the mesenchymal subtype of GBM on transplantation, consistent with a potential astroglial origin for mesenchymal GBM. Sequence analysis of transposon insertion sites from tumors and immortalized cells identified more than 200 frequently mutated genes, including human GBM-associated genes, such as Met and Nf1, and made it possible to discriminate between genes that function during astroglial immortalization vs. later stages of tumor development. We also functionally validated five GBM candidate genes using a previously undescribed high-throughput method. Finally, we show that even clonally related tumors derived from the same immortalized line have acquired distinct combinations of genetic alterations during tumor development, suggesting that tumor formation in this model system involves competition among genetically variant cells, which is similar to the Darwinian evolutionary processes now thought to generate many human cancers. This mutagenesis strategy is faster and simpler than conventional transposon screens and can potentially be applied to any tissue stem/progenitor cells that can be grown and differentiated in vitro.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Combination of a ptgs2 inhibitor and an epidermal growth factor receptor-signaling inhibitor prevents tumorigenesis of oligodendrocyte lineage-derived glioma-initiating cells.Stem Cells. 2011 Apr;29(4):590-9. doi: 10.1002/stem.618. Stem Cells. 2011. PMID: 21360625
-
Glioblastoma extracellular vesicles induce the tumour-promoting transformation of neural stem cells.Cancer Lett. 2019 Dec 1;466:1-12. doi: 10.1016/j.canlet.2019.09.004. Epub 2019 Sep 12. Cancer Lett. 2019. PMID: 31521694
-
Constitutive Notch2 signaling in neural stem cells promotes tumorigenic features and astroglial lineage entry.Cell Death Dis. 2012 Jun 21;3(6):e325. doi: 10.1038/cddis.2012.65. Cell Death Dis. 2012. PMID: 22717580 Free PMC article.
-
New aspects of glioblastoma multiforme revealed by similarities between neural and glioblastoma stem cells.Cell Biol Toxicol. 2018 Dec;34(6):425-440. doi: 10.1007/s10565-017-9420-y. Epub 2018 Jan 31. Cell Biol Toxicol. 2018. PMID: 29383547 Review.
-
Signals that regulate the oncogenic fate of neural stem cells and progenitors.Exp Neurol. 2014 Oct;260:56-68. doi: 10.1016/j.expneurol.2013.01.027. Epub 2013 Jan 31. Exp Neurol. 2014. PMID: 23376224 Free PMC article. Review.
Cited by
-
Single-cell transcriptomic analysis identifies downregulated phosphodiesterase 8B as a novel oncogene in IDH-mutant glioma.Front Immunol. 2024 Jun 26;15:1427200. doi: 10.3389/fimmu.2024.1427200. eCollection 2024. Front Immunol. 2024. PMID: 38989284 Free PMC article.
-
Sleeping Beauty transposon insertional mutagenesis based mouse models for cancer gene discovery.Curr Opin Genet Dev. 2015 Feb;30:66-72. doi: 10.1016/j.gde.2015.04.007. Epub 2015 Jun 4. Curr Opin Genet Dev. 2015. PMID: 26051241 Free PMC article. Review.
-
Cancer gene discovery: exploiting insertional mutagenesis.Mol Cancer Res. 2013 Oct;11(10):1141-58. doi: 10.1158/1541-7786.MCR-13-0244. Epub 2013 Aug 8. Mol Cancer Res. 2013. PMID: 23928056 Free PMC article. Review.
-
Glioblastoma Stem Cells Microenvironment: The Paracrine Roles of the Niche in Drug and Radioresistance.Stem Cells Int. 2016;2016:6809105. doi: 10.1155/2016/6809105. Epub 2016 Jan 6. Stem Cells Int. 2016. PMID: 26880981 Free PMC article. Review.
-
Spontaneous development of intratumoral heterogeneity in a transposon-induced mouse model of glioma.Cancer Sci. 2018 May;109(5):1513-1523. doi: 10.1111/cas.13579. Epub 2018 Apr 15. Cancer Sci. 2018. PMID: 29575648 Free PMC article.
References
-
- Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. - PubMed
-
- Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. - PubMed
-
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

