Tumor-associated mutations in O⁶ -methylguanine DNA-methyltransferase (MGMT) reduce DNA repair functionality
- PMID: 23065697
- PMCID: PMC3720794
- DOI: 10.1002/mc.21964
Tumor-associated mutations in O⁶ -methylguanine DNA-methyltransferase (MGMT) reduce DNA repair functionality
Abstract
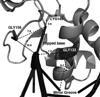
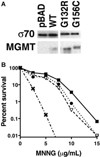
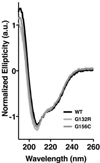
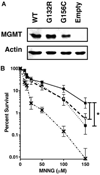
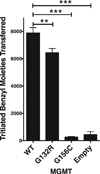
MGMT is the primary vehicle for cellular removal of alkyl lesions from the O-6 position of guanine and the O-4 position of thymine. While key to the maintenance of genomic integrity, MGMT also removes damage induced by alkylating chemotherapies, inhibiting the efficacy of cancer treatment. Germline variants of human MGMT are well-characterized, but somatic variants found in tumors were, prior to this work, uncharacterized. We found that MGMT G132R, from a human esophageal tumor, and MGMT G156C, from a human colorectal cancer cell line, are unable to rescue methyltransferase-deficient Escherichia coli as well as wild type (WT) human MGMT after treatment with a methylating agent. Using pre-steady state kinetics, we biochemically characterized these variants as having a reduced rate constant. G132R binds DNA containing an O⁶ -methylguanine lesion half as tightly as WT MGMT, while G156C has a 40-fold decrease in binding affinity for the same damaged DNA versus WT. Mammalian cells expressing either G132R or G156C are more sensitive to methylating agents than mammalian cells expressing WT MGMT. G132R is slightly resistant to O⁶ -benzylguanine, an inhibitor of MGMT in clinical trials, while G156C is almost completely resistant to this inhibitor. The impared functionality of expressed variants G132R and G156C suggests that the presence of somatic variants of MGMT in a tumor could impact chemotherapeutic outcomes.
Keywords: DNA repair; MNNG; O(6)-alkylguanine DNA alkyltransferase; O(6)-benzyguanine; O-(6)-methylguanine.
© 2013 Wiley Periodicals, Inc.
Figures

Similar articles
-
Lipoic acid inhibits the DNA repair protein O 6-methylguanine-DNA methyltransferase (MGMT) and triggers its depletion in colorectal cancer cells with concomitant autophagy induction.Carcinogenesis. 2015 Aug;36(8):817-31. doi: 10.1093/carcin/bgv070. Epub 2015 May 21. Carcinogenesis. 2015. PMID: 25998848
-
Transfection of a human glioblastoma cell line with liver-type glutaminase (LGA) down-regulates the expression of DNA-repair gene MGMT and sensitizes the cells to alkylating agents.J Neurochem. 2012 Nov;123(3):428-36. doi: 10.1111/j.1471-4159.2012.07917.x. Epub 2012 Sep 21. J Neurochem. 2012. PMID: 22888977
-
MGMT inhibition suppresses survivin expression in pancreatic cancer.Pancreas. 2015 May;44(4):626-35. doi: 10.1097/MPA.0000000000000299. Pancreas. 2015. PMID: 25875800
-
MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents.DNA Repair (Amst). 2007 Aug 1;6(8):1079-99. doi: 10.1016/j.dnarep.2007.03.008. Epub 2007 May 7. DNA Repair (Amst). 2007. PMID: 17485253 Review.
-
MGMT: a personal perspective.DNA Repair (Amst). 2007 Aug 1;6(8):1064-70. doi: 10.1016/j.dnarep.2007.03.007. Epub 2007 May 7. DNA Repair (Amst). 2007. PMID: 17482889 Free PMC article. Review.
Cited by
-
Probing altered enzyme activity in the biochemical characterization of cancer.Biosci Rep. 2022 Feb 25;42(2):BSR20212002. doi: 10.1042/BSR20212002. Biosci Rep. 2022. PMID: 35048115 Free PMC article. Review.
-
Non-bulky Lesions in Human DNA: the Ways of Formation, Repair, and Replication.Acta Naturae. 2017 Jul-Sep;9(3):12-26. Acta Naturae. 2017. PMID: 29104772 Free PMC article.
-
In silico profiling and structural insights of zinc metal ion on O6-methylguanine methyl transferase and its interactions using molecular dynamics approach.J Mol Model. 2021 Jan 17;27(2):40. doi: 10.1007/s00894-020-04631-x. J Mol Model. 2021. PMID: 33454889
-
PARP1-MGMT complex underpins pathway crosstalk in O6-methylguanine repair.J Hematol Oncol. 2022 Oct 14;15(1):146. doi: 10.1186/s13045-022-01367-4. J Hematol Oncol. 2022. PMID: 36242092 Free PMC article.
-
MGMT testing--the challenges for biomarker-based glioma treatment.Nat Rev Neurol. 2014 Jul;10(7):372-85. doi: 10.1038/nrneurol.2014.100. Epub 2014 Jun 10. Nat Rev Neurol. 2014. PMID: 24912512 Review.
References
-
- Margison G, Povey A, Santibanez-Koref M. Genetic variation at the human MGMT locus and its biological consequences. Curr Pharmacogen. 2006;4:133–144.
-
- Drablos F, Feyzi E, Aas PA, et al. Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. - PubMed
-
- Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. - PubMed
-
- Pegg AE, Kangula S, Loktionova NA. O6-alkylguanine-DNA alkyltransferase. Chem Carcinog: Curr Cancer Res. 2011:321–343.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

