Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses
- PMID: 23218769
- PMCID: PMC3596458
- DOI: 10.1016/j.smim.2012.11.001
Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses
Abstract
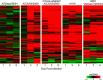
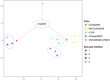
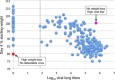
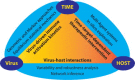
Influenza virus research has recently undergone a shift from a virus-centric perspective to one that embraces the full spectrum of virus-host interactions and cellular signaling events that determine disease outcome. This change has been brought about by the increasing use and expanding scope of high-throughput molecular profiling and computational biology, which together fuel discovery in systems biology. In this review, we show how these approaches have revealed an uncontrolled inflammatory response as a contributor to the extreme virulence of the 1918 pandemic and avian H5N1 viruses, and how this response differs from that induced by the 2009 H1N1 viruses responsible for the most recent influenza pandemic. We also discuss how new animal models, such as the Collaborative Cross mouse systems genetics platform, are key to the necessary systematic investigation of the impact of host genetics on infection outcome, how genome-wide RNAi screens have identified hundreds of cellular factors involved in viral replication, and how systems biology approaches are making possible the rational design of new drugs and vaccines against an ever-evolving respiratory virus.
Keywords: Computational biology; Genomics; Inflammation; Influenza virus; Interferon; Systems biology.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Functional Evolution of the 2009 Pandemic H1N1 Influenza Virus NS1 and PA in Humans.J Virol. 2018 Sep 12;92(19):e01206-18. doi: 10.1128/JVI.01206-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021892 Free PMC article.
-
Dynamic gene expression analysis in a H1N1 influenza virus mouse pneumonia model.Virus Genes. 2017 Jun;53(3):357-366. doi: 10.1007/s11262-017-1438-y. Epub 2017 Feb 27. Virus Genes. 2017. PMID: 28243843
-
2009 pandemic H1N1 influenza A virus strains display differential pathogenicity in C57BL/6J but not BALB/c mice.Virulence. 2011 Nov-Dec;2(6):563-6. doi: 10.4161/viru.2.6.18148. Epub 2011 Nov 1. Virulence. 2011. PMID: 22030859
-
Innate immune evasion strategies of influenza viruses.Future Microbiol. 2010 Jan;5(1):23-41. doi: 10.2217/fmb.09.108. Future Microbiol. 2010. PMID: 20020828 Free PMC article. Review.
-
Pathogenicity and virulence of influenza.Virulence. 2023 Dec;14(1):2223057. doi: 10.1080/21505594.2023.2223057. Virulence. 2023. PMID: 37339323 Free PMC article. Review.
Cited by
-
Induction and Subversion of Human Protective Immunity: Contrasting Influenza and Respiratory Syncytial Virus.Front Immunol. 2018 Mar 2;9:323. doi: 10.3389/fimmu.2018.00323. eCollection 2018. Front Immunol. 2018. PMID: 29552008 Free PMC article. Review.
-
Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses.mBio. 2014 May 20;5(3):e01174-14. doi: 10.1128/mBio.01174-14. mBio. 2014. PMID: 24846384 Free PMC article.
-
Vaccinomics, adversomics, and the immune response network theory: individualized vaccinology in the 21st century.Semin Immunol. 2013 Apr;25(2):89-103. doi: 10.1016/j.smim.2013.04.007. Epub 2013 Jun 5. Semin Immunol. 2013. PMID: 23755893 Free PMC article. Review.
-
Current Insights into the Host Immune Response to Respiratory Viral Infections.Adv Exp Med Biol. 2021;1313:59-83. doi: 10.1007/978-3-030-67452-6_4. Adv Exp Med Biol. 2021. PMID: 34661891
-
Antiviral innate immunity through the lens of systems biology.Virus Res. 2016 Jun 15;218:10-7. doi: 10.1016/j.virusres.2015.11.024. Epub 2015 Dec 3. Virus Res. 2016. PMID: 26657882 Free PMC article. Review.
References
-
- Hale B.G., Randall R.E., Ortin J., Jackson D. The multifunctional NS1 protein of influenza A viruses. Journal of General Virology. 2008;89:2359–2376. - PubMed
-
- Pizzorno A., Abed Y., Boivin G. Influenza drug resistance. Seminars in Respiratory and Critical Care Medicine. 2011;32:409–422. - PubMed
-
- Geiss G.K., Salvatore M., Tumpey T.M., Carter V.S., Wang X., Basler C.F. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10736–10741. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

