Systems biology, bioinformatics, and biomarkers in neuropsychiatry
- PMID: 23269912
- PMCID: PMC3529307
- DOI: 10.3389/fnins.2012.00187
Systems biology, bioinformatics, and biomarkers in neuropsychiatry
Abstract
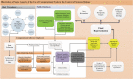
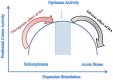
Although neuropsychiatric (NP) disorders are among the top causes of disability worldwide with enormous financial costs, they can still be viewed as part of the most complex disorders that are of unknown etiology and incomprehensible pathophysiology. The complexity of NP disorders arises from their etiologic heterogeneity and the concurrent influence of environmental and genetic factors. In addition, the absence of rigid boundaries between the normal and diseased state, the remarkable overlap of symptoms among conditions, the high inter-individual and inter-population variations, and the absence of discriminative molecular and/or imaging biomarkers for these diseases makes difficult an accurate diagnosis. Along with the complexity of NP disorders, the practice of psychiatry suffers from a "top-down" method that relied on symptom checklists. Although checklist diagnoses cost less in terms of time and money, they are less accurate than a comprehensive assessment. Thus, reliable and objective diagnostic tools such as biomarkers are needed that can detect and discriminate among NP disorders. The real promise in understanding the pathophysiology of NP disorders lies in bringing back psychiatry to its biological basis in a systemic approach which is needed given the NP disorders' complexity to understand their normal functioning and response to perturbation. This approach is implemented in the systems biology discipline that enables the discovery of disease-specific NP biomarkers for diagnosis and therapeutics. Systems biology involves the use of sophisticated computer software "omics"-based discovery tools and advanced performance computational techniques in order to understand the behavior of biological systems and identify diagnostic and prognostic biomarkers specific for NP disorders together with new _targets of therapeutics. In this review, we try to shed light on the need of systems biology, bioinformatics, and biomarkers in neuropsychiatry, and illustrate how the knowledge gained through these methodologies can be translated into clinical use providing clinicians with improved ability to diagnose, manage, and treat NP patients.
Keywords: autism; bioinformatics; biomarkers; data mining; omics; proteomics; psychiatry; systems biology.
Figures

Similar articles
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Bioinformatics approach to understanding interacting pathways in neuropsychiatric disorders.Methods Mol Biol. 2014;1168:157-72. doi: 10.1007/978-1-4939-0847-9_9. Methods Mol Biol. 2014. PMID: 24870135 Review.
-
American Medical Society for Sports Medicine position statement: concussion in sport.Br J Sports Med. 2013 Jan;47(1):15-26. doi: 10.1136/bjsports-2012-091941. Br J Sports Med. 2013. PMID: 23243113 Review.
-
Evidence Brief: The Quality of Care Provided by Advanced Practice Nurses [Internet].Washington (DC): Department of Veterans Affairs (US); 2014 Sep. Washington (DC): Department of Veterans Affairs (US); 2014 Sep. PMID: 27606392 Free Books & Documents. Review.
-
Systems Biology Applications to Decipher Mechanisms and Novel Biomarkers in CNS Trauma.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 30. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 30. PMID: 26269919 Free Books & Documents. Review.
Cited by
-
Antibody profiling of bipolar disorder using Escherichia coli proteome microarrays.Mol Cell Proteomics. 2015 Mar;14(3):510-8. doi: 10.1074/mcp.M114.045930. Epub 2014 Dec 24. Mol Cell Proteomics. 2015. PMID: 25540388 Free PMC article.
-
Exploring Biomarkers for Alzheimer's Disease.J Clin Diagn Res. 2016 Jul;10(7):KE01-6. doi: 10.7860/JCDR/2016/18828.8166. Epub 2016 Jul 1. J Clin Diagn Res. 2016. PMID: 27630867 Free PMC article. Review.
-
Toward a Global Roadmap for Precision Medicine in Psychiatry: Challenges and Opportunities.OMICS. 2016 Oct;20(10):557-564. doi: 10.1089/omi.2016.0110. Epub 2016 Sep 16. OMICS. 2016. PMID: 27636104 Free PMC article. Review.
-
Identification of Biomarkers in Neuropsychiatric Disorders Based on Systems Biology and Epigenetics.Front Genet. 2019 Oct 11;10:985. doi: 10.3389/fgene.2019.00985. eCollection 2019. Front Genet. 2019. PMID: 31681422 Free PMC article. Review.
-
Model-based optimization approaches for precision medicine: A case study in presynaptic dopamine overactivity.PLoS One. 2017 Jun 14;12(6):e0179575. doi: 10.1371/journal.pone.0179575. eCollection 2017. PLoS One. 2017. PMID: 28614410 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

