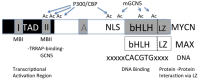
MYCN and the epigenome
- PMID: 23373009
- PMCID: PMC3555505
- DOI: 10.3389/fonc.2013.00001
MYCN and the epigenome
Abstract
It is well known that Neuroblastoma (NB) patients whose tumors have an undifferentiated histology and a transcriptome enriched in cell cycle genes have a worse prognosis. This contrasts with the good prognoses of patients whose tumors have histologic evidence of differentiation and a transcriptome enriched in differentiation genes. Tumor cell lines from poor prognosis, high-risk patients contain a number of genetic alterations, including amplification of MYCN, 1pLOH, and unbalanced 11q or gains of Chr 17 and 7, and exhibit uncontrolled growth and an undifferentiated phenotype in in vitro culture. Yet treatment of such NB cell lines with retinoic acid results in growth control and induction of differentiation. This indicates that the signaling pathways that regulate cell growth and differentiation are not functionally lost but dysregulated. Agents such as retinoic acid normalize the signaling pathways and impose growth control and induction of differentiation. Recent studies in embryonic stem cells indicate that polycomb repressor complex proteins (PRC1 and PRC2) play a major role in regulating stem cell lineage specification and coordinating the shift from a transcriptome that supports self-renewal or growth to one that specifies lineage and controls growth. We have shown that in NB, the PRC2 complex is elevated in undifferentiated NB tumors and functions to suppress a number of tumor suppressor genes. This study will review the role of MYC genes in regulating the epigenome in normal development and explore how this role may be altered during tumorigenesis.
Keywords: EZH2; HAT; HDAC; MYCN; RNA polymerase II; epigenetics; nucleosome; transcriptional activation.
Figures


Similar articles
-
Incorporating genomic, transcriptomic and clinical data: a prognostic and stem cell-like MYC and PRC imbalance in high-risk neuroblastoma.BMC Syst Biol. 2017 Oct 3;11(Suppl 5):92. doi: 10.1186/s12918-017-0466-5. BMC Syst Biol. 2017. PMID: 28984200 Free PMC article.
-
EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR.Cancer Res. 2012 Jan 1;72(1):315-24. doi: 10.1158/0008-5472.CAN-11-0961. Epub 2011 Nov 8. Cancer Res. 2012. PMID: 22068036 Free PMC article.
-
Bmi1 is a MYCN _target gene that regulates tumorigenesis through repression of KIF1Bbeta and TSLC1 in neuroblastoma.Oncogene. 2010 May 6;29(18):2681-90. doi: 10.1038/onc.2010.22. Epub 2010 Mar 1. Oncogene. 2010. PMID: 20190806
-
Proliferation and Survival of Embryonic Sympathetic Neuroblasts by MYCN and Activated ALK Signaling.J Neurosci. 2016 Oct 5;36(40):10425-10439. doi: 10.1523/JNEUROSCI.0183-16.2016. J Neurosci. 2016. PMID: 27707976 Free PMC article.
-
The MYCN oncogene and differentiation in neuroblastoma.Semin Cancer Biol. 2011 Oct;21(4):256-66. doi: 10.1016/j.semcancer.2011.08.001. Epub 2011 Aug 9. Semin Cancer Biol. 2011. PMID: 21849159 Review.
Cited by
-
The role of histone demethylase KDM4B in Myc signaling in neuroblastoma.J Natl Cancer Inst. 2015 Apr 29;107(6):djv080. doi: 10.1093/jnci/djv080. Print 2015 Jun. J Natl Cancer Inst. 2015. PMID: 25925418 Free PMC article.
-
Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma.J Clin Invest. 2020 Nov 2;130(11):5875-5892. doi: 10.1172/JCI134132. J Clin Invest. 2020. PMID: 33016930 Free PMC article.
-
MYCN Silencing by RNAi Induces Neurogenesis and Suppresses Proliferation in Models of Neuroblastoma with Resistance to Retinoic Acid.Nucleic Acid Ther. 2020 Aug;30(4):237-248. doi: 10.1089/nat.2019.0831. Epub 2020 Apr 2. Nucleic Acid Ther. 2020. PMID: 32240058 Free PMC article.
-
Meeting the Challenge of _targeting Cancer Stem Cells.Front Cell Dev Biol. 2019 Feb 18;7:16. doi: 10.3389/fcell.2019.00016. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 30834247 Free PMC article. Review.
-
A Chemo-Genomic Approach Identifies Diverse Epigenetic Therapeutic Vulnerabilities in MYCN-Amplified Neuroblastoma.Front Cell Dev Biol. 2021 Apr 21;9:612518. doi: 10.3389/fcell.2021.612518. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33968920 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

