Endoplasmic reticulum stress sensing in the unfolded protein response
- PMID: 23388626
- PMCID: PMC3578356
- DOI: 10.1101/cshperspect.a013169
Endoplasmic reticulum stress sensing in the unfolded protein response
Abstract
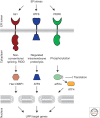
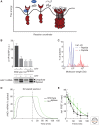
Secretory and transmembrane proteins enter the endoplasmic reticulum (ER) as unfolded proteins and exit as either folded proteins in transit to their _target organelles or as misfolded proteins _targeted for degradation. The unfolded protein response (UPR) maintains the protein-folding homeostasis within the ER, ensuring that the protein-folding capacity of the ER meets the load of client proteins. Activation of the UPR depends on three ER stress sensor proteins, Ire1, PERK, and ATF6. Although the consequences of activation are well understood, how these sensors detect ER stress remains unclear. Recent evidence suggests that yeast Ire1 directly binds to unfolded proteins, which induces its oligomerization and activation. BiP dissociation from Ire1 regulates this oligomeric equilibrium, ultimately modulating Ire1's sensitivity and duration of activation. The mechanistic principles of ER stress sensing are the focus of this review.
Figures


Comment in
-
Endoplasmic reticulum stress and fungal pathogenesis converge.Virulence. 2014 Feb 15;5(2):331-3. doi: 10.4161/viru.28051. Epub 2014 Feb 6. Virulence. 2014. PMID: 24504109 Free PMC article. No abstract available.
Similar articles
-
Endoplasmic reticulum stress and fungal pathogenesis converge.Virulence. 2014 Feb 15;5(2):331-3. doi: 10.4161/viru.28051. Epub 2014 Feb 6. Virulence. 2014. PMID: 24504109 Free PMC article. No abstract available.
-
Molecular signal networks and regulating mechanisms of the unfolded protein response.J Zhejiang Univ Sci B. 2017 Jan.;18(1):1-14. doi: 10.1631/jzus.B1600043. J Zhejiang Univ Sci B. 2017. PMID: 28070992 Free PMC article. Review.
-
Imaging of single cell responses to ER stress indicates that the relative dynamics of IRE1/XBP1 and PERK/ATF4 signalling rather than a switch between signalling branches determine cell survival.Cell Death Differ. 2015 Sep;22(9):1502-16. doi: 10.1038/cdd.2014.241. Epub 2015 Jan 30. Cell Death Differ. 2015. PMID: 25633195 Free PMC article.
-
UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor.Nat Struct Mol Biol. 2019 Nov;26(11):1053-1062. doi: 10.1038/s41594-019-0324-9. Epub 2019 Nov 6. Nat Struct Mol Biol. 2019. PMID: 31695187 Free PMC article.
-
The Unfolded Protein Response and Cell Fate Control.Mol Cell. 2018 Jan 18;69(2):169-181. doi: 10.1016/j.molcel.2017.06.017. Epub 2017 Nov 5. Mol Cell. 2018. PMID: 29107536 Review.
Cited by
-
Chaperone BiP controls ER stress sensor Ire1 through interactions with its oligomers.Life Sci Alliance. 2024 Aug 5;7(10):e202402702. doi: 10.26508/lsa.202402702. Print 2024 Oct. Life Sci Alliance. 2024. PMID: 39103227 Free PMC article.
-
Gut Microbiome Critically Impacts PCB-induced Changes in Metabolic Fingerprints and the Hepatic Transcriptome in Mice.Toxicol Sci. 2020 Sep 1;177(1):168-187. doi: 10.1093/toxsci/kfaa090. Toxicol Sci. 2020. PMID: 32544245 Free PMC article.
-
Regulation of cell-non-autonomous proteostasis in metazoans.Essays Biochem. 2016 Oct 15;60(2):133-142. doi: 10.1042/EBC20160006. Essays Biochem. 2016. PMID: 27744329 Free PMC article. Review.
-
Heat shock protein 47 confers chemoresistance on pancreatic cancer cells by interacting with calreticulin and IRE1α.Cancer Sci. 2021 Jul;112(7):2803-2820. doi: 10.1111/cas.14976. Epub 2021 Jun 9. Cancer Sci. 2021. PMID: 34109710 Free PMC article.
-
Caveolin-1 suppresses tumor formation through the inhibition of the unfolded protein response.Cell Death Dis. 2020 Aug 3;11(8):648. doi: 10.1038/s41419-020-02792-4. Cell Death Dis. 2020. PMID: 32811828 Free PMC article.
References
-
- Abravaya K, Myers MP, Murphy SP, Morimoto RI 1992. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev 6: 1153–1164 - PubMed
-
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K 2008. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct 33: 75–89 - PubMed
-
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and insigs. J Biol Chem 279: 52772–52780 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
