Dissecting the pathways that destabilize mutant p53: the proteasome or autophagy?
- PMID: 23466706
- PMCID: PMC3646859
- DOI: 10.4161/cc.24128
Dissecting the pathways that destabilize mutant p53: the proteasome or autophagy?
Abstract
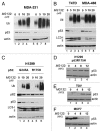
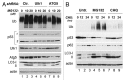
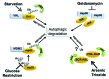
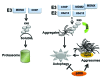
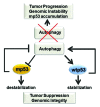
One fundamental feature of mutant forms of p53 consists in their accumulation at high levels in tumors. At least in the case of neomorphic p53 mutations, which acquire oncogenic activity, stabilization is a driving force for tumor progression. It is well documented that p53 mutants are resistant to proteasome-dependent degradation compared with wild-type p53, but the exact identity of the pathways that affect mutant p53 stability is still debated. We have recently shown that macroautophagy (autophagy) provides a route for p53 mutant degradation during restriction of glucose. Here we further show that in basal conditions of growth, inhibition of autophagy with chemical inhibitors or by downregulation of the essential autophagic genes ATG1/Ulk1, Beclin-1 or ATG5, results in p53 mutant stabilization. Conversely, overexpression of Beclin-1 or ATG1/Ulk1 leads to p53 mutant depletion. Furthermore, we found that in many cell lines, prolonged inhibition of the proteasome does not stabilize mutant p53 but leads to its autophagic-mediated degradation. Therefore, we conclude that autophagy is a key mechanism for regulating the stability of several p53 mutants. We discuss plausible mechanisms involved in this newly identified degradation pathway as well as the possible role played by autophagy during tumor evolution driven by mutant p53.
Keywords: autophagy; cancer; mutant; mutations; p53; proteasome; ubiquitin tumor.
Figures

Similar articles
-
The GST-BHMT assay reveals a distinct mechanism underlying proteasome inhibition-induced macroautophagy in mammalian cells.Autophagy. 2015;11(5):812-32. doi: 10.1080/15548627.2015.1034402. Autophagy. 2015. PMID: 25984893 Free PMC article.
-
Phospholipase D-mediated autophagic regulation is a potential _target for cancer therapy.Cell Death Differ. 2014 Apr;21(4):533-46. doi: 10.1038/cdd.2013.174. Epub 2013 Dec 6. Cell Death Differ. 2014. PMID: 24317201 Free PMC article.
-
Radiation induces autophagic cell death via the p53/DRAM signaling pathway in breast cancer cells.Oncol Rep. 2016 Jun;35(6):3639-47. doi: 10.3892/or.2016.4752. Epub 2016 Apr 19. Oncol Rep. 2016. PMID: 27109777
-
Regulation of p53 stability as a therapeutic strategy for cancer.Biochem Pharmacol. 2021 Mar;185:114407. doi: 10.1016/j.bcp.2021.114407. Epub 2021 Jan 7. Biochem Pharmacol. 2021. PMID: 33421376 Review.
-
Molecules and their functions in autophagy.Exp Mol Med. 2012 Feb 29;44(2):73-80. doi: 10.3858/emm.2012.44.2.029. Exp Mol Med. 2012. PMID: 22257882 Free PMC article. Review.
Cited by
-
Small chaperons and autophagy protected neurons from necrotic cell death.Sci Rep. 2017 Jul 18;7(1):5650. doi: 10.1038/s41598-017-05995-6. Sci Rep. 2017. PMID: 28720827 Free PMC article.
-
_targeting Autophagy in ALK-Associated Cancers.Cancers (Basel). 2017 Nov 27;9(12):161. doi: 10.3390/cancers9120161. Cancers (Basel). 2017. PMID: 29186933 Free PMC article. Review.
-
Combination of Proteasome and Histone Deacetylase Inhibitors Overcomes the Impact of Gain-of-Function p53 Mutations.Dis Markers. 2018 Dec 17;2018:3810108. doi: 10.1155/2018/3810108. eCollection 2018. Dis Markers. 2018. PMID: 30647797 Free PMC article.
-
Metabolic stress controls mutant p53 R248Q stability in acute myeloid leukemia cells.Sci Rep. 2019 Apr 4;9(1):5637. doi: 10.1038/s41598-019-42220-y. Sci Rep. 2019. PMID: 30948782 Free PMC article.
-
p53-Autophagy-Metastasis Link.Cancers (Basel). 2018 May 18;10(5):148. doi: 10.3390/cancers10050148. Cancers (Basel). 2018. PMID: 29783720 Free PMC article. Review.
References
-
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
