Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value
- PMID: 23700391
- PMCID: PMC3660251
- DOI: 10.1371/journal.pmed.1001453
Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value
Abstract
Background: Colon cancer (CC) pathological staging fails to accurately predict recurrence, and to date, no gene expression signature has proven reliable for prognosis stratification in clinical practice, perhaps because CC is a heterogeneous disease. The aim of this study was to establish a comprehensive molecular classification of CC based on mRNA expression profile analyses.
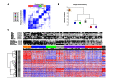
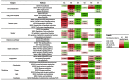
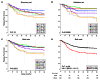
Methods and findings: Fresh-frozen primary tumor samples from a large multicenter cohort of 750 patients with stage I to IV CC who underwent surgery between 1987 and 2007 in seven centers were characterized for common DNA alterations, including BRAF, KRAS, and TP53 mutations, CpG island methylator phenotype, mismatch repair status, and chromosomal instability status, and were screened with whole genome and transcriptome arrays. 566 samples fulfilled RNA quality requirements. Unsupervised consensus hierarchical clustering applied to gene expression data from a discovery subset of 443 CC samples identified six molecular subtypes. These subtypes were associated with distinct clinicopathological characteristics, molecular alterations, specific enrichments of supervised gene expression signatures (stem cell phenotype-like, normal-like, serrated CC phenotype-like), and deregulated signaling pathways. Based on their main biological characteristics, we distinguished a deficient mismatch repair subtype, a KRAS mutant subtype, a cancer stem cell subtype, and three chromosomal instability subtypes, including one associated with down-regulated immune pathways, one with up-regulation of the Wnt pathway, and one displaying a normal-like gene expression profile. The classification was validated in the remaining 123 samples plus an independent set of 1,058 CC samples, including eight public datasets. Furthermore, prognosis was analyzed in the subset of stage II-III CC samples. The subtypes C4 and C6, but not the subtypes C1, C2, C3, and C5, were independently associated with shorter relapse-free survival, even after adjusting for age, sex, stage, and the emerging prognostic classifier Oncotype DX Colon Cancer Assay recurrence score (hazard ratio 1.5, 95% CI 1.1-2.1, p = 0.0097). However, a limitation of this study is that information on tumor grade and number of nodes examined was not available.
Conclusions: We describe the first, to our knowledge, robust transcriptome-based classification of CC that improves the current disease stratification based on clinicopathological variables and common DNA markers. The biological relevance of these subtypes is illustrated by significant differences in prognosis. This analysis provides possibilities for improving prognostic models and therapeutic strategies. In conclusion, we report a new classification of CC into six molecular subtypes that arise through distinct biological pathways.
Conflict of interest statement
YP gave expert advice to Coloplast (France) for less than 1,000 euros last year. The other authors declare that no competing interests exist.
Figures

Similar articles
-
Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes.Gastroenterology. 2015 Jan;148(1):88-99. doi: 10.1053/j.gastro.2014.09.041. Epub 2014 Oct 8. Gastroenterology. 2015. PMID: 25305506 Free PMC article. Clinical Trial.
-
Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis.Lancet Oncol. 2013 Dec;14(13):1295-306. doi: 10.1016/S1470-2045(13)70491-1. Epub 2013 Nov 13. Lancet Oncol. 2013. PMID: 24239208
-
Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer.J Clin Oncol. 2012 Apr 20;30(12):1288-95. doi: 10.1200/JCO.2011.39.5814. Epub 2012 Mar 5. J Clin Oncol. 2012. PMID: 22393095
-
What We Know About Stage II and III Colon Cancer: It's Still Not Enough._target Oncol. 2017 Jun;12(3):265-275. doi: 10.1007/s11523-017-0494-5. _target Oncol. 2017. PMID: 28504299 Free PMC article. Review.
-
Evolving role of gene expression signatures as biomarkers in early-stage colon cancer.J Gastrointest Cancer. 2014 Dec;45(4):399-404. doi: 10.1007/s12029-014-9634-7. J Gastrointest Cancer. 2014. PMID: 24989938 Review.
Cited by
-
Clinical Value of Consensus Molecular Subtypes in Colorectal Cancer: A Systematic Review and Meta-Analysis.J Natl Cancer Inst. 2022 Apr 11;114(4):503-516. doi: 10.1093/jnci/djab106. J Natl Cancer Inst. 2022. PMID: 34077519 Free PMC article.
-
Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis.Br J Cancer. 2015 Sep 1;113(5):763-72. doi: 10.1038/bjc.2015.260. Epub 2015 Jul 21. Br J Cancer. 2015. PMID: 26196185 Free PMC article.
-
Identification of lncRNA-mRNA network linking ferroptosis and immune infiltration to colon adenocarcinoma suppression.Heliyon. 2024 Jun 27;10(13):e33738. doi: 10.1016/j.heliyon.2024.e33738. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39050439 Free PMC article.
-
The significance of combining VEGFA, FLT1, and KDR expressions in colon cancer patient prognosis and predicting response to bevacizumab.Onco _targets Ther. 2015 Apr 15;8:835-43. doi: 10.2147/OTT.S80518. eCollection 2015. Onco _targets Ther. 2015. PMID: 25926745 Free PMC article.
-
Transcriptome analysis reveals a reprogramming energy metabolism-related signature to improve prognosis in colon cancer.PeerJ. 2020 Jul 7;8:e9458. doi: 10.7717/peerj.9458. eCollection 2020. PeerJ. 2020. PMID: 32704448 Free PMC article.
References
-
- Greenlee RT, Murray T, Bolden S, Wingo PA (2000) Cancer statistics, 2000. CA Cancer J Clin 50: 7–33. - PubMed
-
- American Joint Committee on Cancer (1997) AJCC cancer staging manual, 5th edition. Philadelphia: Lippincott-Raven.
-
- Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23: 609–618. - PubMed
-
- Hutchins G, Southward K, Handley K, Magill L, Beaumont C, et al. (2011) Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol 29: 1261–1270. - PubMed
-
- Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, et al. (2004) Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol 22: 1564–1571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

