The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit
- PMID: 23708518
- PMCID: PMC3735711
- DOI: 10.4161/cc.25035
The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit
Abstract
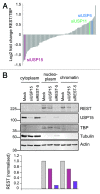
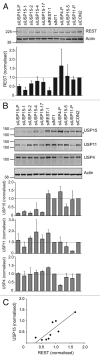
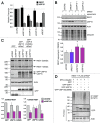
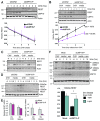
Reversible ubiquitylation of proteins contributes to their integrity, abundance and activity. The RE1-silencing transcription factor (REST) plays key physiological roles and is dysregulated in a spectrum of disease. It is rapidly turned over and is phosphorylated, polyubiquitylated and degraded en masse during neuronal differentiation and cell cycle progression. Through siRNA screening we identified the deubiquitylase USP15 as a key regulator of cellular REST. Both antagonism of REST polyubiquitylation and rescue of endogenous REST levels are dependent on the deubiquitylase activity of USP15. However, USP15 depletion does not destabilize pre-existing REST, but rather specifically impairs de novo REST synthesis. Indeed, we find that a small fraction of endogenous USP15 is associated with polysomes. In accordance with these findings, USP15 does not antagonize the degradation of phosphorylated REST at mitosis. Instead it is required for the rapid accumulation of newly synthesized REST on mitotic exit, thus playing a key role in its cell cycle oscillations. Importantly, this study reveals a novel role for a DUB in specifically promoting new protein synthesis.
Keywords: G1; NRSF; cell cycle; co-translational; deubiquitination; post-translational modification; protein degradation; ubiquitin specific peptidase 15.
Figures
Comment in
-
DUBs "found in translation": USP15 controls stability of newly synthesized REST.Cell Cycle. 2013 Aug 15;12(16):2536-7. doi: 10.4161/cc.25844. Epub 2013 Jul 30. Cell Cycle. 2013. PMID: 23907157 Free PMC article. No abstract available.
Similar articles
-
The deubiquitylase USP15 regulates topoisomerase II alpha to maintain genome integrity.Oncogene. 2018 Apr;37(17):2326-2342. doi: 10.1038/s41388-017-0092-0. Epub 2018 Feb 12. Oncogene. 2018. PMID: 29429988 Free PMC article.
-
Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells.Nat Cell Biol. 2011 Feb;13(2):142-52. doi: 10.1038/ncb2153. Epub 2011 Jan 23. Nat Cell Biol. 2011. PMID: 21258371 Free PMC article.
-
The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation.Oncogene. 2013 Mar 28;32(13):1691-701. doi: 10.1038/onc.2012.182. Epub 2012 Jun 4. Oncogene. 2013. PMID: 22665064 Free PMC article.
-
Impressionist portraits of mitotic exit: APC/C, K11-linked ubiquitin chains and Cezanne.Cell Cycle. 2019 Mar-Apr;18(6-7):652-660. doi: 10.1080/15384101.2019.1593646. Epub 2019 Mar 28. Cell Cycle. 2019. PMID: 30874463 Free PMC article. Review.
-
The Multifaceted Roles of USP15 in Signal Transduction.Int J Mol Sci. 2021 Apr 29;22(9):4728. doi: 10.3390/ijms22094728. Int J Mol Sci. 2021. PMID: 33946990 Free PMC article. Review.
Cited by
-
Ubiquitination of RORγt at Lysine 446 Limits Th17 Differentiation by Controlling Coactivator Recruitment.J Immunol. 2016 Aug 15;197(4):1148-58. doi: 10.4049/jimmunol.1600548. Epub 2016 Jul 18. J Immunol. 2016. PMID: 27430721 Free PMC article.
-
USP15 in Cancer and Other Diseases: From Diverse Functionsto Therapeutic _targets.Biomedicines. 2022 Feb 17;10(2):474. doi: 10.3390/biomedicines10020474. Biomedicines. 2022. PMID: 35203682 Free PMC article. Review.
-
REST alleviates neurotoxic prion peptide-induced synaptic abnormalities, neurofibrillary degeneration and neuronal death partially via LRP6-mediated Wnt-β-catenin signaling.Onco_target. 2016 Mar 15;7(11):12035-52. doi: 10.18632/onco_target.7640. Onco_target. 2016. PMID: 26919115 Free PMC article.
-
A flexible microfluidic system for single-cell transcriptome profiling elucidates phased transcriptional regulators of cell cycle.Sci Rep. 2021 Apr 12;11(1):7918. doi: 10.1038/s41598-021-86070-z. Sci Rep. 2021. PMID: 33846365 Free PMC article.
-
DUBbing Cancer: Deubiquitylating Enzymes Involved in Epigenetics, DNA Damage and the Cell Cycle As Therapeutic _targets.Front Genet. 2016 Jul 28;7:133. doi: 10.3389/fgene.2016.00133. eCollection 2016. Front Genet. 2016. PMID: 27516771 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
