Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry
- PMID: 24101521
- PMCID: PMC3808593
- DOI: 10.1073/pnas.1312374110
Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry
Abstract
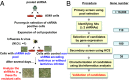
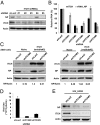
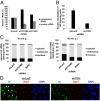
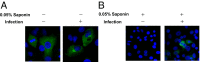
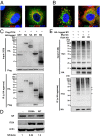
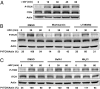
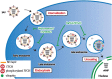
Influenza viruses, like other viruses, rely on host factors to support their life cycle as viral proteins usually "hijack," or collaborate with, cellular proteins to execute their functions. Identification and understanding of these factors can increase the knowledge of molecular mechanisms manipulated by the viruses and facilitate development of antiviral drugs. To this end, we developed a unique genome-wide pooled shRNA screen to search for cellular factors important for influenza A virus (IAV) replication. We identified an E3 ubiquitin ligase, Itch, as an essential factor for an early step in the viral life cycle. In Itch knockdown cells, the incorporation of viral ribonucleoprotein complex into endosomes was normal, but its subsequent release from endosomes and transport to the nucleus was retarded. In addition, upon virus infection, Itch was phosphorylated and recruited to the endosomes, where virus particles were located. Furthermore, Itch interacted with viral M1 protein and ubiquitinated M1 protein. Collectively, our findings unravel a critical role of Itch in mediating IAV release from the endosome and offer insights into the mechanism for IAV uncoating during virus entry. These findings also highlight the feasibility of pooled RNAi screening for exploring the cellular cofactors of lytic viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
CypA Regulates AIP4-Mediated M1 Ubiquitination of Influenza A Virus.Virol Sin. 2018 Oct;33(5):440-448. doi: 10.1007/s12250-018-0058-6. Epub 2018 Oct 16. Virol Sin. 2018. PMID: 30328013 Free PMC article.
-
Cathepsin W Is Required for Escape of Influenza A Virus from Late Endosomes.mBio. 2015 Jun 9;6(3):e00297. doi: 10.1128/mBio.00297-15. mBio. 2015. PMID: 26060270 Free PMC article.
-
Interaction of NS2 with AIMP2 facilitates the switch from ubiquitination to SUMOylation of M1 in influenza A virus-infected cells.J Virol. 2015 Jan;89(1):300-11. doi: 10.1128/JVI.02170-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320310 Free PMC article.
-
Ubiquitin in Influenza Virus Entry and Innate Immunity.Viruses. 2016 Oct 24;8(10):293. doi: 10.3390/v8100293. Viruses. 2016. PMID: 27783058 Free PMC article. Review.
-
Entry of influenza A virus into host cells - recent progress and remaining challenges.Curr Opin Virol. 2021 Jun;48:23-29. doi: 10.1016/j.coviro.2021.03.001. Epub 2021 Apr 7. Curr Opin Virol. 2021. PMID: 33838498 Review.
Cited by
-
Valosin-containing protein/p97 plays critical roles in the Japanese encephalitis virus life cycle.J Virol. 2021 May 10;95(11):e02336-20. doi: 10.1128/JVI.02336-20. Epub 2021 Mar 17. J Virol. 2021. PMID: 33731458 Free PMC article.
-
Hepatitis C Virus-Induced ROS/JNK Signaling Pathway Activates the E3 Ubiquitin Ligase Itch to Promote the Release of HCV Particles via Polyubiquitylation of VPS4A.J Virol. 2022 Mar 23;96(6):e0181121. doi: 10.1128/JVI.01811-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35044214 Free PMC article.
-
Sequence-Specific Features of Short Double-Strand, Blunt-End RNAs Have RIG-I- and Type 1 Interferon-Dependent or -Independent Anti-Viral Effects.Viruses. 2022 Jun 28;14(7):1407. doi: 10.3390/v14071407. Viruses. 2022. PMID: 35891387 Free PMC article.
-
Ubiquitination of the Cytoplasmic Domain of Influenza A Virus M2 Protein Is Crucial for Production of Infectious Virus Particles.J Virol. 2018 Jan 30;92(4):e01972-17. doi: 10.1128/JVI.01972-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29167343 Free PMC article.
-
H5N1 infection impairs the alveolar epithelial barrier through intercellular junction proteins via Itch-mediated proteasomal degradation.Commun Biol. 2022 Mar 1;5(1):186. doi: 10.1038/s42003-022-03131-3. Commun Biol. 2022. PMID: 35233032 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

