Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes
- PMID: 24103892
- PMCID: PMC3814599
- DOI: 10.3390/v5102483
Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes
Abstract
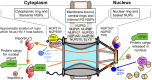
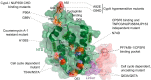
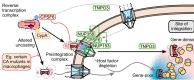
Retroviruses integrate their reverse transcribed genomes into host cell chromosomes as an obligate step in virus replication. The nuclear envelope separates the chromosomes from the cell cytoplasm during interphase, and different retroviral groups deal with this physical barrier in different ways. Gammaretroviruses are dependent on the passage of _target cells through mitosis, where they are believed to access chromosomes when the nuclear envelope dissolves for cell division. Contrastingly, lentiviruses such as HIV-1 infect non-dividing cells, and are believed to enter the nucleus by passing through the nuclear pore complex. While numerous virally encoded elements have been proposed to be involved in HIV-1 nuclear import, recent evidence has highlighted the importance of HIV-1 capsid. Furthermore, capsid was found to be responsible for the viral requirement of various nuclear transport proteins, including transportin 3 and nucleoporins NUP153 and NUP358, during infection. In this review, we describe our current understanding of retroviral nuclear import, with emphasis on recent developments on the role of the HIV-1 capsid protein.
Figures

Similar articles
-
Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells.J Virol. 2018 Sep 12;92(19):e00648-18. doi: 10.1128/JVI.00648-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997211 Free PMC article.
-
HIV Capsid and Integration _targeting.Viruses. 2021 Jan 18;13(1):125. doi: 10.3390/v13010125. Viruses. 2021. PMID: 33477441 Free PMC article. Review.
-
Factors that mold the nuclear landscape of HIV-1 integration.Nucleic Acids Res. 2021 Jan 25;49(2):621-635. doi: 10.1093/nar/gkaa1207. Nucleic Acids Res. 2021. PMID: 33337475 Free PMC article. Review.
-
The nuclear pore complex: a new dynamic in HIV-1 replication.Nucleus. 2010 Jan-Feb;1(1):18-22. doi: 10.4161/nucl.1.1.10571. Nucleus. 2010. PMID: 21327100 Free PMC article.
-
Nuclear Import of HIV-1.Viruses. 2021 Nov 8;13(11):2242. doi: 10.3390/v13112242. Viruses. 2021. PMID: 34835048 Free PMC article. Review.
Cited by
-
Retroviral DNA Transposition: Themes and Variations.Microbiol Spectr. 2014 Dec;2(5):MDNA300052014. doi: 10.1128/microbiolspec.MDNA3-0005-2014. Microbiol Spectr. 2014. PMID: 25844274 Free PMC article.
-
Recent Advances in HIV-1 Gag Inhibitor Design and Development.Molecules. 2020 Apr 7;25(7):1687. doi: 10.3390/molecules25071687. Molecules. 2020. PMID: 32272714 Free PMC article. Review.
-
A critical role for alternative polyadenylation factor CPSF6 in _targeting HIV-1 integration to transcriptionally active chromatin.Proc Natl Acad Sci U S A. 2016 Feb 23;113(8):E1054-63. doi: 10.1073/pnas.1524213113. Epub 2016 Feb 8. Proc Natl Acad Sci U S A. 2016. PMID: 26858452 Free PMC article.
-
Control of yeast retrotransposons mediated through nucleoporin evolution.PLoS Genet. 2018 Apr 25;14(4):e1007325. doi: 10.1371/journal.pgen.1007325. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29694349 Free PMC article.
-
The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration.Nucleic Acids Res. 2018 Apr 20;46(7):3552-3578. doi: 10.1093/nar/gky109. Nucleic Acids Res. 2018. PMID: 29514267 Free PMC article.
References
-
- Gartner S., Markovits P., Markovitz D.M., Kaplan M.H., Gallo R.C., Popovic M. The Role of Mononuclear Phagocytes in HTLV–III/LAV Infection. Science. 1986;233:215–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

