PGC7 suppresses TET3 for protecting DNA methylation
- PMID: 24322296
- PMCID: PMC3950671
- DOI: 10.1093/nar/gkt1261
PGC7 suppresses TET3 for protecting DNA methylation
Abstract
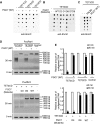
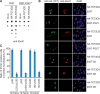
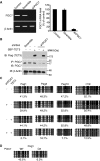
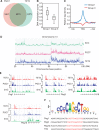
Ten-eleven translocation (TET) family enzymes convert 5-methylcytosine to 5-hydroxylmethylcytosine. However, the molecular mechanism that regulates this biological process is not clear. Here, we show the evidence that PGC7 (also known as Dppa3 or Stella) interacts with TET2 and TET3 both in vitro and in vivo to suppress the enzymatic activity of TET2 and TET3. Moreover, lacking PGC7 induces the loss of DNA methylation at imprinting loci. Genome-wide analysis of PGC7 reveals a consensus DNA motif that is recognized by PGC7. The CpG islands surrounding the PGC7-binding motifs are hypermethylated. Taken together, our study demonstrates a molecular mechanism by which PGC7 protects DNA methylation from TET family enzyme-dependent oxidation.
Figures

Similar articles
-
TET proteins and 5-methylcytosine oxidation in hematological cancers.Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review.
-
PGC7, H3K9me2 and Tet3: regulators of DNA methylation in zygotes.Cell Res. 2013 Jan;23(1):6-9. doi: 10.1038/cr.2012.117. Epub 2012 Aug 7. Cell Res. 2013. PMID: 22868271 Free PMC article.
-
PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos.Nature. 2012 Jun 3;486(7403):415-9. doi: 10.1038/nature11093. Nature. 2012. PMID: 22722204
-
Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX.Nature. 2013 May 2;497(7447):122-6. doi: 10.1038/nature12052. Epub 2013 Apr 7. Nature. 2013. PMID: 23563267 Free PMC article.
-
Epigenetic Function of TET Family, 5-Methylcytosine, and 5-Hydroxymethylcytosine in Hematologic Malignancies.Oncol Res Treat. 2019;42(6):309-318. doi: 10.1159/000498947. Epub 2019 May 3. Oncol Res Treat. 2019. PMID: 31055566 Review.
Cited by
-
EHMT2 and SETDB1 protect the maternal pronucleus from 5mC oxidation.Proc Natl Acad Sci U S A. 2019 May 28;116(22):10834-10841. doi: 10.1073/pnas.1819946116. Epub 2019 May 14. Proc Natl Acad Sci U S A. 2019. PMID: 31088968 Free PMC article.
-
DPPA3 prevents cytosine hydroxymethylation of the maternal pronucleus and is required for normal development in bovine embryos.Epigenetics. 2014 Sep;9(9):1271-9. doi: 10.4161/epi.32087. Epub 2014 Aug 1. Epigenetics. 2014. PMID: 25147917 Free PMC article.
-
DPPA3 facilitates genome-wide DNA demethylation in mouse primordial germ cells.BMC Genomics. 2024 Apr 5;25(1):344. doi: 10.1186/s12864-024-10192-7. BMC Genomics. 2024. PMID: 38580899 Free PMC article.
-
TET proteins and 5-methylcytosine oxidation in hematological cancers.Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review.
-
Potential Roles of Intrinsic Disorder in Maternal-Effect Proteins Involved in the Maintenance of DNA Methylation.Int J Mol Sci. 2017 Sep 4;18(9):1898. doi: 10.3390/ijms18091898. Int J Mol Sci. 2017. PMID: 28869544 Free PMC article.
References
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. - PubMed
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007;19:266–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

