AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data
- PMID: 24389661
- PMCID: PMC6280749
- DOI: 10.1093/bioinformatics/btt759
AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data
Abstract
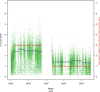
Motivation: Detection and quantification of the absolute DNA copy number alterations in tumor cells is challenging because the DNA specimen is extracted from a mixture of tumor and normal stromal cells. Estimates of tumor purity and ploidy are necessary to correctly infer copy number, and ploidy may itself be a prognostic factor in cancer progression. As deep sequencing of the exome or genome has become routine for characterization of tumor samples, in this work, we aim to develop a simple and robust algorithm to infer purity, ploidy and absolute copy numbers in whole numbers for tumor cells from sequencing data.
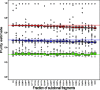
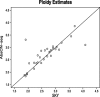
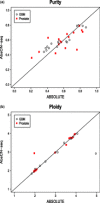
Results: A simulation study shows that estimates have reasonable accuracy, and that the algorithm is robust against the presence of segmentation errors and subclonal populations. We validated our algorithm against a panel of cell lines with experimentally determined ploidy. We also compared our algorithm with the well-established single-nucleotide polymorphism array-based method called ABSOLUTE on three sets of tumors of different types. Our method had good performance on these four benchmark datasets for both purity and ploidy estimates, and may offer a simple solution to copy number alteration quantification for cancer sequencing projects.
Availability and implementation: The R package absCNseq is available from http://biostats.mcc.ucsd.edu/files/absCNseq_1.0.tar.gz CONTACT: kmesser@ucsd.edu Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

Similar articles
-
Accurity: accurate tumor purity and ploidy inference from tumor-normal WGS data by jointly modelling somatic copy number alterations and heterozygous germline single-nucleotide-variants.Bioinformatics. 2018 Jun 15;34(12):2004-2011. doi: 10.1093/bioinformatics/bty043. Bioinformatics. 2018. PMID: 29385401 Free PMC article.
-
Deconvolving tumor purity and ploidy by integrating copy number alterations and loss of heterozygosity.Bioinformatics. 2014 Aug 1;30(15):2121-9. doi: 10.1093/bioinformatics/btu174. Epub 2014 Apr 2. Bioinformatics. 2014. PMID: 24695406 Free PMC article.
-
Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data.Ann Oncol. 2015 Jan;26(1):64-70. doi: 10.1093/annonc/mdu479. Epub 2014 Oct 15. Ann Oncol. 2015. PMID: 25319062 Free PMC article.
-
Using SAAS-CNV to Detect and Characterize Somatic Copy Number Alterations in Cancer Genomes from Next Generation Sequencing and SNP Array Data.Methods Mol Biol. 2018;1833:29-47. doi: 10.1007/978-1-4939-8666-8_2. Methods Mol Biol. 2018. PMID: 30039361 Review.
-
Methods for copy number aberration detection from single-cell DNA-sequencing data.Genome Biol. 2020 Aug 17;21(1):208. doi: 10.1186/s13059-020-02119-8. Genome Biol. 2020. PMID: 32807205 Free PMC article. Review.
Cited by
-
Tissue-specific cell-free DNA degradation quantifies circulating tumor DNA burden.Nat Commun. 2021 Apr 13;12(1):2229. doi: 10.1038/s41467-021-22463-y. Nat Commun. 2021. PMID: 33850132 Free PMC article.
-
ENVE: a novel computational framework characterizes copy-number mutational landscapes in colorectal cancers from African American patients.Genome Med. 2015 Jul 20;7(1):69. doi: 10.1186/s13073-015-0192-9. eCollection 2015. Genome Med. 2015. PMID: 26269717 Free PMC article.
-
Classification of HER2-negative breast cancers by ERBB2 copy number alteration status reveals molecular differences associated with chromosome 17 gene aberrations.Ther Adv Med Oncol. 2023 Oct 31;15:17588359231206259. doi: 10.1177/17588359231206259. eCollection 2023. Ther Adv Med Oncol. 2023. PMID: 37920257 Free PMC article.
-
Global copy number profiling of cancer genomes.Bioinformatics. 2016 Mar 15;32(6):926-8. doi: 10.1093/bioinformatics/btv676. Epub 2015 Nov 16. Bioinformatics. 2016. PMID: 26576652 Free PMC article.
-
Mutational Profiling Can Establish Clonal or Independent Origin in Synchronous Bilateral Breast and Other Tumors.PLoS One. 2015 Nov 10;10(11):e0142487. doi: 10.1371/journal.pone.0142487. eCollection 2015. PLoS One. 2015. PMID: 26554380 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

