Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine
- PMID: 24522915
- PMCID: PMC3993788
- DOI: 10.1128/JVI.03805-13
Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine
Abstract
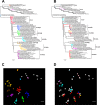
Swine influenza A virus is an endemic and economically important pathogen in pigs, with the potential to infect other host species. The hemagglutinin (HA) protein is the primary _target of protective immune responses and the major component in swine influenza A vaccines. However, as a result of antigenic drift, vaccine strains must be regularly updated to reflect currently circulating strains. Characterizing the cross-reactivity between strains in pigs and seasonal influenza virus strains in humans is also important in assessing the relative risk of interspecies transmission of viruses from one host population to the other. Hemagglutination inhibition (HI) assay data for swine and human H3N2 viruses were used with antigenic cartography to quantify the antigenic differences among H3N2 viruses isolated from pigs in the United States from 1998 to 2013 and the relative cross-reactivity between these viruses and current human seasonal influenza A virus strains. Two primary antigenic clusters were found circulating in the pig population, but with enough diversity within and between the clusters to suggest updates in vaccine strains are needed. We identified single amino acid substitutions that are likely responsible for antigenic differences between the two primary antigenic clusters and between each antigenic cluster and outliers. The antigenic distance between current seasonal influenza virus H3 strains in humans and those endemic in swine suggests that population immunity may not prevent the introduction of human viruses into pigs, and possibly vice versa, reinforcing the need to monitor and prepare for potential incursions.
Importance: Influenza A virus (IAV) is an important pathogen in pigs and humans. The hemagglutinin (HA) protein is the primary _target of protective immune responses and the major _target of vaccines. However, vaccine strains must be updated to reflect current strains. Characterizing the differences between seasonal IAV in humans and swine IAV is important in assessing the relative risk of interspecies transmission of viruses. We found two primary antigenic clusters of H3N2 in the U.S. pig population, with enough diversity to suggest updates in swine vaccine strains are needed. We identified changes in the HA protein that are likely responsible for these differences and that may be useful in predicting when vaccines need to be updated. The difference between human H3N2 viruses and those in swine is enough that population immunity is unlikely to prevent new introductions of human IAV into pigs or vice versa, reinforcing the need to monitor and prepare for potential introductions.
Figures

Similar articles
-
Evolution and Antigenic Advancement of N2 Neuraminidase of Swine Influenza A Viruses Circulating in the United States following Two Separate Introductions from Human Seasonal Viruses.J Virol. 2021 Sep 27;95(20):e0063221. doi: 10.1128/JVI.00632-21. Epub 2021 Aug 11. J Virol. 2021. PMID: 34379513 Free PMC article.
-
The Molecular Determinants of Antibody Recognition and Antigenic Drift in the H3 Hemagglutinin of Swine Influenza A Virus.J Virol. 2016 Aug 26;90(18):8266-80. doi: 10.1128/JVI.01002-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27384658 Free PMC article.
-
Novel Reassortant Human-Like H3N2 and H3N1 Influenza A Viruses Detected in Pigs Are Virulent and Antigenically Distinct from Swine Viruses Endemic to the United States.J Virol. 2015 Nov;89(22):11213-22. doi: 10.1128/JVI.01675-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311895 Free PMC article.
-
H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation.Hum Vaccin Immunother. 2018;14(8):1840-1847. doi: 10.1080/21645515.2018.1462639. Epub 2018 May 14. Hum Vaccin Immunother. 2018. PMID: 29641358 Free PMC article. Review.
-
Swine Influenza A Viruses and the Tangled Relationship with Humans.Cold Spring Harb Perspect Med. 2021 Mar 1;11(3):a038737. doi: 10.1101/cshperspect.a038737. Cold Spring Harb Perspect Med. 2021. PMID: 31988203 Free PMC article. Review.
Cited by
-
Human-Origin Influenza A(H3N2) Reassortant Viruses in Swine, Southeast Mexico.Emerg Infect Dis. 2019 Apr;25(4):691-700. doi: 10.3201/eid2504.180779. Epub 2019 Apr 17. Emerg Infect Dis. 2019. PMID: 30730827 Free PMC article.
-
Genetic and antigenic evolution of H1 swine influenza A viruses isolated in Belgium and the Netherlands from 2014 through 2019.Sci Rep. 2021 May 28;11(1):11276. doi: 10.1038/s41598-021-90512-z. Sci Rep. 2021. PMID: 34050216 Free PMC article.
-
Evolution and Antigenic Advancement of N2 Neuraminidase of Swine Influenza A Viruses Circulating in the United States following Two Separate Introductions from Human Seasonal Viruses.J Virol. 2021 Sep 27;95(20):e0063221. doi: 10.1128/JVI.00632-21. Epub 2021 Aug 11. J Virol. 2021. PMID: 34379513 Free PMC article.
-
Heterologous prime-boost vaccination with H3N2 influenza viruses of swine favors cross-clade antibody responses and protection.NPJ Vaccines. 2017;2:11. doi: 10.1038/s41541-017-0012-x. Epub 2017 Apr 20. NPJ Vaccines. 2017. PMID: 29250437 Free PMC article.
-
The global antigenic diversity of swine influenza A viruses.Elife. 2016 Apr 15;5:e12217. doi: 10.7554/eLife.12217. Elife. 2016. PMID: 27113719 Free PMC article.
References
-
- Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus ADME. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822. 10.1128/JVI.79.5.2814-2822.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

