Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy
- PMID: 24695223
- PMCID: PMC4180099
- DOI: 10.1038/nature13148
Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy
Abstract
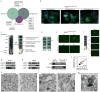
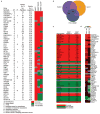
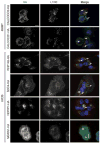
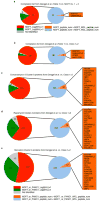
Autophagy, the process by which proteins and organelles are sequestered in double-membrane structures called autophagosomes and delivered to lysosomes for degradation, is critical in diseases such as cancer and neurodegeneration. Much of our understanding of this process has emerged from analysis of bulk cytoplasmic autophagy, but our understanding of how specific cargo, including organelles, proteins or intracellular pathogens, are _targeted for selective autophagy is limited. Here we use quantitative proteomics to identify a cohort of novel and known autophagosome-enriched proteins in human cells, including cargo receptors. Like known cargo receptors, nuclear receptor coactivator 4 (NCOA4) was highly enriched in autophagosomes, and associated with ATG8 proteins that recruit cargo-receptor complexes into autophagosomes. Unbiased identification of NCOA4-associated proteins revealed ferritin heavy and light chains, components of an iron-filled cage structure that protects cells from reactive iron species but is degraded via autophagy to release iron through an unknown mechanism. We found that delivery of ferritin to lysosomes required NCOA4, and an inability of NCOA4-deficient cells to degrade ferritin led to decreased bioavailable intracellular iron. This work identifies NCOA4 as a selective cargo receptor for autophagic turnover of ferritin (ferritinophagy), which is critical for iron homeostasis, and provides a resource for further dissection of autophagosomal cargo-receptor connectivity.
Figures






Comment in
-
Identifying specific receptors for cargo-mediated autophagy.Cell Res. 2014 Jul;24(7):783-4. doi: 10.1038/cr.2014.56. Epub 2014 May 6. Cell Res. 2014. PMID: 24797431 Free PMC article.
Similar articles
-
Identifying specific receptors for cargo-mediated autophagy.Cell Res. 2014 Jul;24(7):783-4. doi: 10.1038/cr.2014.56. Epub 2014 May 6. Cell Res. 2014. PMID: 24797431 Free PMC article.
-
Expression and characterization of the ferritin binding domain of Nuclear Receptor Coactivator-4 (NCOA4).Biochim Biophys Acta Gen Subj. 2017 Nov;1861(11 Pt A):2710-2716. doi: 10.1016/j.bbagen.2017.07.015. Epub 2017 Jul 25. Biochim Biophys Acta Gen Subj. 2017. PMID: 28754384
-
Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis.Elife. 2015 Oct 5;4:e10308. doi: 10.7554/eLife.10308. Elife. 2015. PMID: 26436293 Free PMC article.
-
The Role of NCOA4-Mediated Ferritinophagy in Ferroptosis.Adv Exp Med Biol. 2021;1301:41-57. doi: 10.1007/978-3-030-62026-4_4. Adv Exp Med Biol. 2021. PMID: 34370287 Review.
-
Ferritinophagy: Molecular mechanisms and role in disease.Pathol Res Pract. 2024 Oct;262:155553. doi: 10.1016/j.prp.2024.155553. Epub 2024 Aug 22. Pathol Res Pract. 2024. PMID: 39180800 Review.
Cited by
-
Ciclopirox olamine induces ferritinophagy and reduces cyst burden in polycystic kidney disease.JCI Insight. 2021 Mar 30;6(8):e141299. doi: 10.1172/jci.insight.141299. JCI Insight. 2021. PMID: 33784251 Free PMC article.
-
Host and microbiota derived extracellular vesicles: Crucial players in iron homeostasis.Front Med (Lausanne). 2022 Oct 13;9:985141. doi: 10.3389/fmed.2022.985141. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36314015 Free PMC article. Review.
-
Genome-wide DNA methylation sequencing reveals the involvement of ferroptosis in hepatotoxicity induced by dietary exposure to food-grade titanium dioxide.Part Fibre Toxicol. 2024 Sep 18;21(1):37. doi: 10.1186/s12989-024-00598-2. Part Fibre Toxicol. 2024. PMID: 39294687 Free PMC article.
-
Potential Roles and Mechanisms of Curcumin and its Derivatives in the Regulation of Ferroptosis.Int J Biol Sci. 2024 Sep 9;20(12):4838-4852. doi: 10.7150/ijbs.90798. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309443 Free PMC article. Review.
-
Quercetin Protects against MPP+/MPTP-Induced Dopaminergic Neuron Death in Parkinson's Disease by Inhibiting Ferroptosis.Oxid Med Cell Longev. 2022 Sep 5;2022:7769355. doi: 10.1155/2022/7769355. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36105483 Free PMC article.
References
Methods Reference
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

