A splice mutation and mRNA decay of EXT2 provoke hereditary multiple exostoses
- PMID: 24728384
- PMCID: PMC3984245
- DOI: 10.1371/journal.pone.0094848
A splice mutation and mRNA decay of EXT2 provoke hereditary multiple exostoses
Abstract
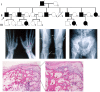
Background: Hereditary multiple exostoses (HME) is an autosomal dominant disease. The classical paradigm of mutation screening seeks to relate alterations in the exostosin glycosyltransferase genes, EXT1 and EXT2, which are responsible for over 70% of HME cases. However, the pathological significance of the majority of these mutations is often unclear.
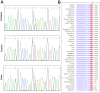
Methods: In a Chinese family with HME, EXT1 and EXT2 genes were screened by direct sequencing. The consequence of a detected mutant was predicted by in silico analysis and confirmed by mRNA analysis. The EXT1 and EXT2 mRNA and protein levels and the HS patterns in the HME patients were compared with those in healthy controls.
Results: A heterozygous transition (c.743+1G>A) in the EXT2 gene, which co-segregated with the HME phenotype in this family, was identified. The G residue at position +1 in intron 4 of EXT2 was predicted to be a 5' donor splice site. The mRNA analysis revealed an alternative transcript with a cryptic splice site 5 bp downstream of the wild-type site, which harbored a premature stop codon. However, the predicted truncated protein was not detected by western blot analysis. Decay of the mutant mRNA was shown by clone sequencing and quantification analysis. The corresponding downregulation of the EXT2 mRNA will contribute to the abnormal EXT1/EXT2 ratio and HS pattern that were detected in the patients with HME.
Conclusion: The heterozygous mutation c.743+1G>A in the EXT2 gene causes HME as a result of abnormal splicing, mRNA decay, and the resulting haploinsufficiency of EXT2.
Conflict of interest statement
Figures


Similar articles
-
A novel splice mutation induces exon skipping of the EXT1 gene in patients with hereditary multiple exostoses.Int J Oncol. 2019 Mar;54(3):859-868. doi: 10.3892/ijo.2019.4688. Epub 2019 Jan 16. Int J Oncol. 2019. PMID: 30664192 Free PMC article.
-
Novel and recurrent mutations in the EXT1 and EXT2 genes in Chinese kindreds with multiple osteochondromas.J Orthop Res. 2013 Sep;31(9):1492-9. doi: 10.1002/jor.22378. Epub 2013 Apr 29. J Orthop Res. 2013. PMID: 23629877
-
Three novel EXT1 and EXT2 gene mutations in Taiwanese patients with multiple exostoses.J Formos Med Assoc. 2006 May;105(5):434-7. doi: 10.1016/s0929-6646(09)60143-1. J Formos Med Assoc. 2006. PMID: 16638657
-
Molecular basis of multiple exostoses: mutations in the EXT1 and EXT2 genes.Hum Mutat. 2000;15(3):220-7. doi: 10.1002/(SICI)1098-1004(200003)15:3<220::AID-HUMU2>3.0.CO;2-K. Hum Mutat. 2000. PMID: 10679937 Review.
-
The pathogenic roles of heparan sulfate deficiency in hereditary multiple exostoses.Matrix Biol. 2018 Oct;71-72:28-39. doi: 10.1016/j.matbio.2017.12.011. Epub 2017 Dec 24. Matrix Biol. 2018. PMID: 29277722 Free PMC article. Review.
Cited by
-
A novel splice mutation induces exon skipping of the EXT1 gene in patients with hereditary multiple exostoses.Int J Oncol. 2019 Mar;54(3):859-868. doi: 10.3892/ijo.2019.4688. Epub 2019 Jan 16. Int J Oncol. 2019. PMID: 30664192 Free PMC article.
-
Identification of a novel EXT2 frameshift mutation in a family with hereditary multiple exostoses by whole-exome sequencing.J Clin Lab Anal. 2021 Sep;35(9):e23968. doi: 10.1002/jcla.23968. Epub 2021 Aug 17. J Clin Lab Anal. 2021. PMID: 34403521 Free PMC article.
-
Association of two synonymous splicing-associated CpG single nucleotide polymorphisms in calpain 10 and solute carrier family 2 member 2 with type 2 diabetes.Biomed Rep. 2017 Feb;6(2):146-158. doi: 10.3892/br.2016.833. Epub 2016 Dec 29. Biomed Rep. 2017. PMID: 28357066 Free PMC article.
-
Exome Analysis Identifies a Novel Compound Heterozygous Alteration in TGM1 Gene Leading to Lamellar Ichthyosis in a Child From Saudi Arabia: Case Presentation.Front Pediatr. 2019 Feb 21;7:44. doi: 10.3389/fped.2019.00044. eCollection 2019. Front Pediatr. 2019. PMID: 30847336 Free PMC article.
-
Donor Splice Site Variant in SLC9A6 Causes Christianson Syndrome in a Lithuanian Family: A Case Report.Medicina (Kaunas). 2022 Feb 26;58(3):351. doi: 10.3390/medicina58030351. Medicina (Kaunas). 2022. PMID: 35334527 Free PMC article.
References
-
- Pannier S, Legeai-Mallet L (2008) Hereditary multiple exostoses and enchondromatosis. Best Practice & Research in Clinical Rheumatology 22: 45–54. - PubMed
-
- Sugiura Y, Sugiura I, Iwata H (1976) HEREDITARY MULTIPLE EXOSTOSIS - DIAPHYSEAL ACLASIS. Japanese Journal of Human Genetics 21: 149–167. - PubMed
-
- Wicklund CL, Pauli RM, Johnston D, Hecht JT (1995) NATURAL-HISTORY STUDY OF HEREDITARY MULTIPLE EXOSTOSES. American Journal of Medical Genetics 55: 43–46. - PubMed
-
- Ahn J, Josefludecke H, Lindow S, Horton WA, Lee B, et al. (1995) CLONING OF THE PUTATIVE TUMOR-SUPPRESSOR GENE FOR HEREDITARY MULTIPLE EXOSTOSES (EXT1). Nature Genetics 11: 137–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

