RAN1 is involved in plant cold resistance and development in rice (Oryza sativa)
- PMID: 24790113
- PMCID: PMC4071843
- DOI: 10.1093/jxb/eru178
RAN1 is involved in plant cold resistance and development in rice (Oryza sativa)
Abstract
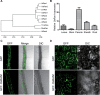
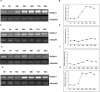
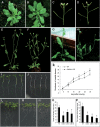
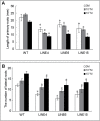
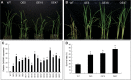
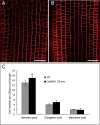
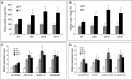
Of the diverse abiotic stresses, low temperature is one of the major limiting factors that lead to a series of morphological, physiological, biochemical, and molecular changes in plants. Ran, an evolutionarily conserved small G-protein family, has been shown to be essential for the nuclear translocation of proteins. It also mediates the regulation of cell cycle progression in mammalian cells. However, little is known about Ran function in rice (Oryza sativa). We report here that Ran gene OsRAN1 is essential for the molecular improvement of rice for cold tolerance. Ran also affects plant morphogenesis in transgenic Arabidopsis thaliana. OsRAN1 is ubiquitously expressed in rice tissues with the highest expression in the spike. The levels of mRNA encoding OsRAN1 were greatly increased by cold and indoleacetic acid treatment rather than by addition of salt and polyethylene glycol. Further, OsRAN1 overexpression in Arabidopsis increased tiller number, and altered root development. OsRAN1 overexpression in rice improves cold tolerance. The levels of cellular free Pro and sugar levels were highly increased in transgenic plants under cold stress. Under cold stress, OsRAN1 maintained cell division and cell cycle progression, and also promoted the formation of an intact nuclear envelope. The results suggest that OsRAN1 protein plays an important role in the regulation of cellular mitosis and the auxin signalling pathway.
Keywords: Cold tolerance; OsRAN1; cell division; intact nuclear envelope.; reduced apical dominance.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


Similar articles
-
OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress.Plant Cell Environ. 2011 Jan;34(1):52-64. doi: 10.1111/j.1365-3040.2010.02225.x. Epub 2010 Oct 4. Plant Cell Environ. 2011. PMID: 20825577
-
Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress.Planta. 2011 Aug;234(2):331-45. doi: 10.1007/s00425-011-1403-2. Epub 2011 Mar 30. Planta. 2011. PMID: 21448719
-
Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes.Plant Physiol. 2009 May;150(1):244-56. doi: 10.1104/pp.108.133454. Epub 2009 Mar 11. Plant Physiol. 2009. PMID: 19279197 Free PMC article.
-
Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin.Plant Physiol. 2006 Jan;140(1):91-101. doi: 10.1104/pp.105.071670. Epub 2005 Dec 16. Plant Physiol. 2006. PMID: 16361516 Free PMC article.
-
Overexpression of a partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress.Plant Sci. 2013 Dec;213:67-78. doi: 10.1016/j.plantsci.2013.08.013. Epub 2013 Sep 7. Plant Sci. 2013. PMID: 24157209
Cited by
-
OsGATA16, a GATA Transcription Factor, Confers Cold Tolerance by Repressing OsWRKY45-1 at the Seedling Stage in Rice.Rice (N Y). 2021 May 12;14(1):42. doi: 10.1186/s12284-021-00485-w. Rice (N Y). 2021. PMID: 33982131 Free PMC article.
-
Emerging role of small GTPases and their interactome in plants to combat abiotic and biotic stress.Protoplasma. 2023 Jul;260(4):1007-1029. doi: 10.1007/s00709-022-01830-6. Epub 2022 Dec 16. Protoplasma. 2023. PMID: 36525153 Review.
-
Molecular characterization and functional analysis of the OsPsbR gene family in rice.Mol Genet Genomics. 2017 Apr;292(2):271-281. doi: 10.1007/s00438-016-1273-1. Epub 2016 Nov 10. Mol Genet Genomics. 2017. PMID: 27832344
-
Genome-wide identification of Ran GTPase family genes from wheat (T. aestivum) and their expression profile during developmental stages and abiotic stress conditions.Funct Integr Genomics. 2021 Mar;21(2):239-250. doi: 10.1007/s10142-021-00773-0. Epub 2021 Feb 20. Funct Integr Genomics. 2021. PMID: 33609188
-
Drought stress tolerance mechanisms and their potential common indicators to salinity, insights from the wild watermelon (Citrullus lanatus): A review.Front Plant Sci. 2023 Feb 6;13:1074395. doi: 10.3389/fpls.2022.1074395. eCollection 2022. Front Plant Sci. 2023. PMID: 36815012 Free PMC article. Review.
References
-
- Abraham E, Rigo G, Szekely G, Nagy R, Koncz C, Szabados L. 2003. Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis . Plant Molecular Biology 51, 363–372 - PubMed
-
- Andaya VC, Tai TH. 2006. Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theoretical and Applied Genetics 113, 467–475 - PubMed
-
- Berleth T, Mattsson J, Hardtke CS. 2000. Vascular continuity and auxin signals. Trends in Plant Science 5, 387–393 - PubMed
-
- Bischoff FR, Ponstingl H. 1991. Catalysis of guanine-nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

