Adventitial fibroblasts induce a distinct proinflammatory/profibrotic macrophage phenotype in pulmonary hypertension
- PMID: 24928992
- PMCID: PMC4100597
- DOI: 10.4049/jimmunol.1303048
Adventitial fibroblasts induce a distinct proinflammatory/profibrotic macrophage phenotype in pulmonary hypertension
Abstract
Macrophage accumulation is not only a characteristic hallmark but is also a critical component of pulmonary artery remodeling associated with pulmonary hypertension (PH). However, the cellular and molecular mechanisms that drive vascular macrophage activation and their functional phenotype remain poorly defined. Using multiple levels of in vivo (bovine and rat models of hypoxia-induced PH, together with human tissue samples) and in vitro (primary mouse, rat, and bovine macrophages, human monocytes, and primary human and bovine fibroblasts) approaches, we observed that adventitial fibroblasts derived from hypertensive pulmonary arteries (bovine and human) regulate macrophage activation. These fibroblasts activate macrophages through paracrine IL-6 and STAT3, HIF1, and C/EBPβ signaling to drive expression of genes previously implicated in chronic inflammation, tissue remodeling, and PH. This distinct fibroblast-activated macrophage phenotype was independent of IL-4/IL-13-STAT6 and TLR-MyD88 signaling. We found that genetic STAT3 haplodeficiency in macrophages attenuated macrophage activation, complete STAT3 deficiency increased macrophage activation through compensatory upregulation of STAT1 signaling, and deficiency in C/EBPβ or HIF1 attenuated fibroblast-driven macrophage activation. These findings challenge the current paradigm of IL-4/IL-13-STAT6-mediated alternative macrophage activation as the sole driver of vascular remodeling in PH, and uncover a cross-talk between adventitial fibroblasts and macrophages in which paracrine IL-6-activated STAT3, HIF1α, and C/EBPβ signaling are critical for macrophage activation and polarization. Thus, _targeting IL-6 signaling in macrophages by completely inhibiting C/EBPβ or HIF1α or by partially inhibiting STAT3 may hold therapeutic value for treatment of PH and other inflammatory conditions characterized by increased IL-6 and absent IL-4/IL-13 signaling.
Copyright © 2014 by The American Association of Immunologists, Inc.
Figures
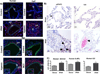
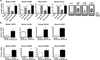
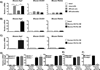
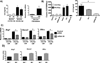
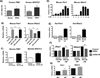
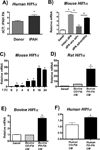
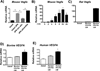

Similar articles
-
Emergence of fibroblasts with a proinflammatory epigenetically altered phenotype in severe hypoxic pulmonary hypertension.J Immunol. 2011 Sep 1;187(5):2711-22. doi: 10.4049/jimmunol.1100479. Epub 2011 Aug 3. J Immunol. 2011. PMID: 21813768 Free PMC article.
-
Microenvironmental Regulation of Macrophage Transcriptomic and Metabolomic Profiles in Pulmonary Hypertension.Front Immunol. 2021 Mar 31;12:640718. doi: 10.3389/fimmu.2021.640718. eCollection 2021. Front Immunol. 2021. PMID: 33868271 Free PMC article.
-
NOX2 deficiency alters macrophage phenotype through an IL-10/STAT3 dependent mechanism: implications for traumatic brain injury.J Neuroinflammation. 2017 Mar 24;14(1):65. doi: 10.1186/s12974-017-0843-4. J Neuroinflammation. 2017. PMID: 28340575 Free PMC article.
-
Metabolic reprogramming and inflammation act in concert to control vascular remodeling in hypoxic pulmonary hypertension.J Appl Physiol (1985). 2015 Nov 15;119(10):1164-72. doi: 10.1152/japplphysiol.00283.2015. Epub 2015 Apr 30. J Appl Physiol (1985). 2015. PMID: 25930027 Free PMC article. Review.
-
The role of inflammation in hypoxic pulmonary hypertension: from cellular mechanisms to clinical phenotypes.Am J Physiol Lung Cell Mol Physiol. 2015 Feb 1;308(3):L229-52. doi: 10.1152/ajplung.00238.2014. Epub 2014 Nov 21. Am J Physiol Lung Cell Mol Physiol. 2015. PMID: 25416383 Free PMC article. Review.
Cited by
-
Inflammation in Pulmonary Arterial Hypertension.Adv Exp Med Biol. 2021;1303:351-372. doi: 10.1007/978-3-030-63046-1_19. Adv Exp Med Biol. 2021. PMID: 33788202 Review.
-
Pharmacological Agents and Potential New Therapies in Pulmonary Arterial Hypertension.Curr Vasc Pharmacol. 2024;22(3):155-170. doi: 10.2174/0115701611266576231211045731. Curr Vasc Pharmacol. 2024. PMID: 38115617 Review.
-
Inositol monophosphatase 1 as a novel interacting partner of RAGE in pulmonary hypertension.Am J Physiol Lung Cell Mol Physiol. 2019 Mar 1;316(3):L428-L444. doi: 10.1152/ajplung.00393.2018. Epub 2019 Jan 3. Am J Physiol Lung Cell Mol Physiol. 2019. PMID: 30604625 Free PMC article.
-
Association Between Circulating CD4+ T Cell Methylation Signatures of Network-Oriented SOCS3 Gene and Hemodynamics in Patients Suffering Pulmonary Arterial Hypertension.J Cardiovasc Transl Res. 2023 Feb;16(1):17-30. doi: 10.1007/s12265-022-10294-1. Epub 2022 Aug 12. J Cardiovasc Transl Res. 2023. PMID: 35960497 Free PMC article.
-
The inflammatory cell landscape in the lungs of patients with idiopathic pulmonary arterial hypertension.Eur Respir J. 2018 Jan 25;51(1):1701214. doi: 10.1183/13993003.01214-2017. Print 2018 Jan. Eur Respir J. 2018. PMID: 29371380 Free PMC article.
References
-
- Morrell NW, Archer SL, Defelice A, Evans S, Fiszman M, Martin T, Saulnier M, Rabinovitch M, Schermuly R, Stewart D, Truebel H, Walker G, Stenmark KR. Anticipated classes of new medications and molecular _targets for pulmonary arterial hypertension. Pulmonary circulation. 2013;3:226–244. - PMC - PubMed
-
- Price LC, Wort SJ, Perros F, Dorfmuller P, Huertas A, Montani D, Cohen-Kaminsky S, Humbert M. Inflammation in pulmonary arterial hypertension. Chest. 2012;141:210–221. - PubMed
-
- Savai R, Pullamsetti SS, Kolbe J, Bieniek E, Voswinckel R, Fink L, Scheed A, Ritter C, Dahal BK, Vater A, Klussmann S, Ghofrani HA, Weissmann N, Klepetko W, Banat GA, Seeger W, Grimminger F, Schermuly RT. Immune and inflammatory cell involvement in the pathology of idiopathic pulmonary arterial hypertension. American journal of respiratory and critical care medicine. 2012;186:897–908. - PubMed
-
- Stenmark KR, Meyrick B, Galie N, Mooi WJ, McMurtry IF. Animal models of pulmonary arterial hypertension: the hope for etiological discovery and pharmacological cure. American journal of physiology. Lung cellular and molecular physiology. 2009;297:L1013–L1032. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

