Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity
- PMID: 24996549
- PMCID: PMC4105547
- DOI: 10.1186/1471-2180-14-182
Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity
Abstract
Background: Candida albicans infections have become increasingly recognised as being biofilm related. Recent studies have shown that there is a relationship between biofilm formation and poor clinical outcomes in patients infected with biofilm proficient strains. Here we have investigated a panel of clinical isolates in an attempt to evaluate their phenotypic and transcriptional properties in an attempt to differentiate and define levels of biofilm formation.
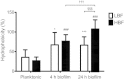
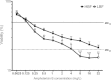
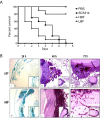
Results: Biofilm formation was shown to be heterogeneous; with isolates being defined as either high or low biofilm formers (LBF and HBF) based on different biomass quantification. These categories could also be differentiated using a cell surface hydrophobicity assay with 24 h biofilms. HBF isolates were more resistance to amphotericin B (AMB) treatment than LBF, but not voriconazole (VRZ). In a Galleria mellonella model of infection HBF mortality was significantly increased in comparison to LBF. Histological analysis of the HBF showed hyphal elements intertwined indicative of the biofilm phenotype. Transcriptional analysis of 23 genes implicated in biofilm formation showed no significant differential expression profiles between LBF and HBF, except for Cdr1 at 4 and 24 h. Cluster analysis showed similar patterns of expression for different functional classes of genes, though correlation analysis of the 4 h biofilms with overall biomass at 24 h showed that 7 genes were correlated with high levels of biofilm, including Als3, Eap1, Cph1, Sap5, Plb1, Cdr1 and Zap1.
Conclusions: Our findings show that biofilm formation is variable amongst C. albicans isolates, and categorising isolates depending on this can be used to predict how pathogenic the isolate will behave clinically. We have shown that looking at individual genes in less informative than looking at multiple genes when trying to categorise isolates at LBF or HBF. These findings are important when developing biofilm-specific diagnostics as these could be used to predict how best to treat patients infected with C. albicans. Further studies are required to evaluate this clinically.
Figures

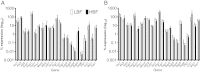
Similar articles
-
Microbiologic and clinical characteristics of biofilm-forming Candida parapsilosis isolates associated with fungaemia and their impact on mortality.Clin Microbiol Infect. 2018 Jul;24(7):771-777. doi: 10.1016/j.cmi.2017.11.005. Epub 2017 Nov 10. Clin Microbiol Infect. 2018. PMID: 29133157
-
Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation.BMC Microbiol. 2014 Dec 5;14:303. doi: 10.1186/s12866-014-0303-6. BMC Microbiol. 2014. PMID: 25476750 Free PMC article.
-
Temporal Profile of Biofilm Formation, Gene Expression and Virulence Analysis in Candida albicans Strains.Mycopathologia. 2017 Apr;182(3-4):285-295. doi: 10.1007/s11046-016-0088-2. Epub 2016 Nov 9. Mycopathologia. 2017. PMID: 27830437
-
Biofilm of Candida albicans: formation, regulation and resistance.J Appl Microbiol. 2021 Jul;131(1):11-22. doi: 10.1111/jam.14949. Epub 2020 Dec 9. J Appl Microbiol. 2021. PMID: 33249681 Review.
-
Prevalence of biofilms in Candida spp. bloodstream infections: A meta-analysis.PLoS One. 2022 Feb 3;17(2):e0263522. doi: 10.1371/journal.pone.0263522. eCollection 2022. PLoS One. 2022. PMID: 35113972 Free PMC article. Review.
Cited by
-
Assessment of Biofilm Formation by Candida albicans Strains Isolated from Hemocultures and Their Role in Pathogenesis in the Zebrafish Model.J Fungi (Basel). 2022 Sep 27;8(10):1014. doi: 10.3390/jof8101014. J Fungi (Basel). 2022. PMID: 36294579 Free PMC article.
-
Distinct Expression Levels of ALS, LIP, and SAP Genes in Candida tropicalis with Diverse Virulent Activities.Front Microbiol. 2016 Jul 29;7:1175. doi: 10.3389/fmicb.2016.01175. eCollection 2016. Front Microbiol. 2016. PMID: 27524980 Free PMC article.
-
Reversal of Azole Resistance in Candida albicans by Human Neutrophil Peptide.Biomedicines. 2023 Feb 10;11(2):513. doi: 10.3390/biomedicines11020513. Biomedicines. 2023. PMID: 36831048 Free PMC article.
-
An intestinal Candida albicans model for monomicrobial and polymicrobial biofilms and effects of hydrolases and the Bgl2 ligand.Vet World. 2022 Apr;15(4):1134-1140. doi: 10.14202/vetworld.2022.1134-1140. Epub 2022 Apr 29. Vet World. 2022. PMID: 35698505 Free PMC article.
-
B-Cell Epitope Mapping from Eight Antigens of Candida albicans to Design a Novel Diagnostic Kit: An Immunoinformatics Approach.Int J Pept Res Ther. 2022;28(4):110. doi: 10.1007/s10989-022-10413-1. Epub 2022 May 27. Int J Pept Res Ther. 2022. PMID: 35669279 Free PMC article.
References
-
- Mensa J, Pitart C, Marco F. Treatment of critically ill patients with candidemia. Int J Antimicrob Agents. 2008;32(Suppl 2):S93–97. - PubMed
-
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. - PubMed
-
- Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med. 2008;59:415–428. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

