The roles of microphthalmia-associated transcription factor and pigmentation in melanoma
- PMID: 25111671
- PMCID: PMC4336945
- DOI: 10.1016/j.abb.2014.07.019
The roles of microphthalmia-associated transcription factor and pigmentation in melanoma
Abstract
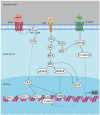
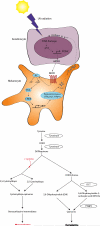
MITF and pigmentation play important roles in both normal melanocyte and transformed melanoma cell biology. MITF is regulated by many pathways and it also regulates many _targets, some of which are still being discovered and functionally validated. MITF is involved in a wide range of processes in melanocytes, including pigment synthesis and lineage survival. Pigmentation itself plays an important role as the interface between genetic and environmental factors that contribute to melanoma.
Keywords: MITF; Melanocyte; Melanoma; Pigmentation.
Figures

Similar articles
-
MITF pathway mutations in melanoma.Pigment Cell Melanoma Res. 2009 Aug;22(4):376-7. doi: 10.1111/j.1755-148X.2009.00599.x. Epub 2009 Jun 25. Pigment Cell Melanoma Res. 2009. PMID: 19558635 No abstract available.
-
Developmental pathways activated in melanocytes and melanoma.Arch Biochem Biophys. 2014 Dec 1;563:13-21. doi: 10.1016/j.abb.2014.07.023. Epub 2014 Aug 8. Arch Biochem Biophys. 2014. PMID: 25109840 Free PMC article. Review.
-
The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation.Oncogene. 2017 Sep 7;36(36):5110-5121. doi: 10.1038/onc.2017.135. Epub 2017 May 8. Oncogene. 2017. PMID: 28481878
-
[Malignant melanoma and the role of the paradoxal protein Microphthalmia transcription factor].Bull Cancer. 2007 Jan;94(1):81-92. Bull Cancer. 2007. PMID: 17237008 Review. French.
-
[The Importance of MITF Signaling Pathway in the Regulation of Proliferation and Invasiveness of Malignant Melanoma].Klin Onkol. 2016 Fall;29(5):347-350. doi: 10.14735/amko2016347. Klin Onkol. 2016. PMID: 27739313 Review. Czech.
Cited by
-
Growth Hormone Upregulates Melanocyte-Inducing Transcription Factor Expression and Activity via JAK2-STAT5 and SRC Signaling in GH Receptor-Positive Human Melanoma.Cancers (Basel). 2019 Sep 12;11(9):1352. doi: 10.3390/cancers11091352. Cancers (Basel). 2019. PMID: 31547367 Free PMC article.
-
Lung cancer deficient in the tumor suppressor GATA4 is sensitive to TGFBR1 inhibition.Nat Commun. 2019 Apr 10;10(1):1665. doi: 10.1038/s41467-019-09295-7. Nat Commun. 2019. PMID: 30971692 Free PMC article.
-
FBXW7 regulates a mitochondrial transcription program by modulating MITF.Pigment Cell Melanoma Res. 2018 Sep;31(5):636-640. doi: 10.1111/pcmr.12704. Epub 2018 Apr 29. Pigment Cell Melanoma Res. 2018. PMID: 29665239 Free PMC article.
-
Context-dependent miR-204 and miR-211 affect the biological properties of amelanotic and melanotic melanoma cells.Onco_target. 2017 Apr 11;8(15):25395-25417. doi: 10.18632/onco_target.15915. Onco_target. 2017. PMID: 28445987 Free PMC article.
-
EdnrB Governs Regenerative Response of Melanocyte Stem Cells by Crosstalk with Wnt Signaling.Cell Rep. 2016 May 10;15(6):1291-302. doi: 10.1016/j.celrep.2016.04.006. Epub 2016 Apr 28. Cell Rep. 2016. PMID: 27134165 Free PMC article.
References
-
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer Journal for Clinicians. 2014;64:9–29. - PubMed
-
- Kudchadkar RR, Gonzalez R, Lewis K. New _targeted therapies in melanoma. Cancer Control. 2013;20:282–288. - PubMed
-
- Melanoma Skin Cancer American Cancer Society. 2013 At < http://www.cancer.org/cancer/skincancer-melanoma/detailedguide/melanoma-...>.
-
- Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nature Reviews Cancer. 2003;3:559–570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

