MIG-10 (Lamellipodin) stabilizes invading cell adhesion to basement membrane and is a negative transcriptional _target of EGL-43 in C. elegans
- PMID: 25148942
- PMCID: PMC4438536
- DOI: 10.1016/j.bbrc.2014.08.049
MIG-10 (Lamellipodin) stabilizes invading cell adhesion to basement membrane and is a negative transcriptional _target of EGL-43 in C. elegans
Abstract
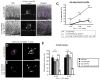
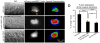
Cell invasion through basement membrane (BM) occurs in many physiological and pathological contexts. MIG-10, the Caenorhabditis elegans Lamellipodin (Lpd), regulates diverse biological processes. Its function and regulation in cell invasive behavior remain unclear. Using anchor cell (AC) invasion in C. elegans as an in vivo invasion model, we have previously found that mig-10's activity is largely outside of UNC-6 (netrin) signaling, a chemical cue directing AC invasion. We have shown that MIG-10 is a _target of the transcription factor FOS-1A and facilitates BM breaching. Combining genetics and imaging analyses, we report that MIG-10 synergizes with UNC-6 to promote AC attachment to the BM, revealing a functional role for MIG-10 in stabilizing AC-BM adhesion. MIG-10 is also required for F-actin accumulation in the absence of UNC-6. Further, we identify mig-10 as a transcriptional _target negatively regulated by EGL-43A (C. elegans Evi-1 proto-oncogene), a transcription factor positively controlled by FOS-1A. The revelation of this negative regulation unmasks an incoherent feedforward circuit existing among fos-1, egl-43 and mig-10. Moreover, our study suggests the functional importance of the negative regulation on mig-10 expression by showing that excessive MIG-10 impairs AC invasion. Thus, we provide new insight into MIG-10's function and its complex transcriptional regulation during cell invasive behavior.
Keywords: Anchor cell; Cell invasion; EGL-43; Lamellipodin; MIG-10.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

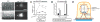
Similar articles
-
MIG-10 (lamellipodin) has netrin-independent functions and is a FOS-1A transcriptional _target during anchor cell invasion in C. elegans.Development. 2014 Mar;141(6):1342-53. doi: 10.1242/dev.102434. Epub 2014 Feb 19. Development. 2014. PMID: 24553288 Free PMC article.
-
UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans.Nat Cell Biol. 2009 Feb;11(2):183-9. doi: 10.1038/ncb1825. Epub 2008 Dec 21. Nat Cell Biol. 2009. PMID: 19098902 Free PMC article.
-
The transcription factor HLH-2/E/Daughterless regulates anchor cell invasion across basement membrane in C. elegans.Dev Biol. 2011 Sep 15;357(2):380-91. doi: 10.1016/j.ydbio.2011.07.012. Epub 2011 Jul 18. Dev Biol. 2011. PMID: 21784067 Free PMC article.
-
Control of the basement membrane and cell migration by ADAMTS proteinases: Lessons from C. elegans genetics.Matrix Biol. 2015 May-Jul;44-46:64-9. doi: 10.1016/j.matbio.2015.01.001. Epub 2015 Jan 14. Matrix Biol. 2015. PMID: 25595837 Review.
-
Control of cell migration during Caenorhabditis elegans development.Curr Opin Cell Biol. 1999 Oct;11(5):608-13. doi: 10.1016/s0955-0674(99)00028-9. Curr Opin Cell Biol. 1999. PMID: 10508660 Review.
Cited by
-
Invading, Leading and Navigating Cells in Caenorhabditis elegans: Insights into Cell Movement in Vivo.Genetics. 2018 Jan;208(1):53-78. doi: 10.1534/genetics.117.300082. Genetics. 2018. PMID: 29301948 Free PMC article. Review.
-
A developmental gene regulatory network for C. elegans anchor cell invasion.Development. 2020 Jan 2;147(1):dev185850. doi: 10.1242/dev.185850. Development. 2020. PMID: 31806663 Free PMC article.
-
The Caenorhabditis elegans homolog of the Evi1 proto-oncogene, egl-43, coordinates G1 cell cycle arrest with pro-invasive gene expression during anchor cell invasion.PLoS Genet. 2020 Mar 23;16(3):e1008470. doi: 10.1371/journal.pgen.1008470. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32203506 Free PMC article.
-
To Divide or Invade: A Look Behind the Scenes of the Proliferation-Invasion Interplay in the Caenorhabditis elegans Anchor Cell.Front Cell Dev Biol. 2021 Jan 6;8:616051. doi: 10.3389/fcell.2020.616051. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33490081 Free PMC article. Review.
-
The SWI/SNF chromatin remodeling assemblies BAF and PBAF differentially regulate cell cycle exit and cellular invasion in vivo.PLoS Genet. 2022 Jan 4;18(1):e1009981. doi: 10.1371/journal.pgen.1009981. eCollection 2022 Jan. PLoS Genet. 2022. PMID: 34982771 Free PMC article.
References
-
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell Migration: Integrating Signals from Front to Back. Science. 2003;302:1704–1709. - PubMed
-
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature reviews Cancer. 2003;3:362–374. - PubMed
-
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

