Structural basis for the recognition of human cytomegalovirus glycoprotein B by a neutralizing human antibody
- PMID: 25299639
- PMCID: PMC4192593
- DOI: 10.1371/journal.ppat.1004377
Structural basis for the recognition of human cytomegalovirus glycoprotein B by a neutralizing human antibody
Abstract
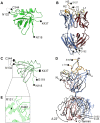
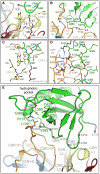
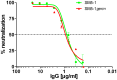
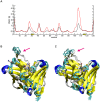
Human cytomegalovirus (HCMV) infections are life-threating to people with a compromised or immature immune system. Upon adhesion, fusion of the virus envelope with the host cell is initiated. In this step, the viral glycoprotein gB is considered to represent the major fusogen. Here, we present for the first time structural data on the binding of an anti-herpes virus antibody and describe the atomic interactions between the antigenic domain Dom-II of HCMV gB and the Fab fragment of the human antibody SM5-1. The crystal structure shows that SM5-1 binds Dom-II almost exclusively via only two CDRs, namely light chain CDR L1 and a 22-residue-long heavy chain CDR H3. Two contiguous segments of Dom-II are _targeted by SM5-1, and the combining site includes a hydrophobic pocket on the Dom-II surface that is only partially filled by CDR H3 residues. SM5-1 belongs to a series of sequence-homologous anti-HCMV gB monoclonal antibodies that were isolated from the same donor at a single time point and that represent different maturation states. Analysis of amino acid substitutions in these antibodies in combination with molecular dynamics simulations show that key contributors to the picomolar affinity of SM5-1 do not directly interact with the antigen but significantly reduce the flexibility of CDR H3 in the bound and unbound state of SM5-1 through intramolecular side chain interactions. Thus, these residues most likely alleviate unfavorable binding entropies associated with extra-long CDR H3s, and this might represent a common strategy during antibody maturation. Models of entire HCMV gB in different conformational states hint that SM5-1 neutralizes HCMV either by blocking the pre- to postfusion transition of gB or by precluding the interaction with additional effectors such as the gH/gL complex.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: NS, HS, THW and MM are inventors on a patent application on the antibody described. This does not alter our adherence to all PLOS policies on sharing data and materials.
Figures



Similar articles
-
A Novel Strain-Specific Neutralizing Epitope on Glycoprotein H of Human Cytomegalovirus.J Virol. 2021 Aug 25;95(18):e0065721. doi: 10.1128/JVI.00657-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34160252 Free PMC article.
-
Recognition of a highly conserved glycoprotein B epitope by a bivalent antibody neutralizing HCMV at a post-attachment step.PLoS Pathog. 2020 Aug 3;16(8):e1008736. doi: 10.1371/journal.ppat.1008736. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32745149 Free PMC article.
-
Monoclonal Antibodies to Different Components of the Human Cytomegalovirus (HCMV) Pentamer gH/gL/pUL128L and Trimer gH/gL/gO as well as Antibodies Elicited during Primary HCMV Infection Prevent Epithelial Cell Syncytium Formation.J Virol. 2016 Jun 24;90(14):6216-6223. doi: 10.1128/JVI.00121-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27122579 Free PMC article.
-
From recognition to execution-the HCMV Pentamer from receptor binding to fusion triggering.Curr Opin Virol. 2018 Aug;31:43-51. doi: 10.1016/j.coviro.2018.05.004. Epub 2018 Jun 1. Curr Opin Virol. 2018. PMID: 29866439 Review.
-
Pathogen at the Gates: Human Cytomegalovirus Entry and Cell Tropism.Viruses. 2018 Dec 11;10(12):704. doi: 10.3390/v10120704. Viruses. 2018. PMID: 30544948 Free PMC article. Review.
Cited by
-
In Vitro Characterization of Human Cytomegalovirus-_targeting Therapeutic Monoclonal Antibodies LJP538 and LJP539.Antimicrob Agents Chemother. 2016 Jul 22;60(8):4961-71. doi: 10.1128/AAC.00382-16. Print 2016 Aug. Antimicrob Agents Chemother. 2016. PMID: 27270290 Free PMC article.
-
A glycoprotein B-neutralizing antibody structure at 2.8 Å uncovers a critical domain for herpesvirus fusion initiation.Nat Commun. 2020 Aug 18;11(1):4141. doi: 10.1038/s41467-020-17911-0. Nat Commun. 2020. PMID: 32811830 Free PMC article.
-
Beacons Contribute Valuable Empirical Information to Theoretical 3-D Aptamer-Peptide Binding.J Fluoresc. 2019 May;29(3):711-717. doi: 10.1007/s10895-019-02380-6. Epub 2019 May 1. J Fluoresc. 2019. PMID: 31044327
-
Crystal Structure of the Human Cytomegalovirus Glycoprotein B.PLoS Pathog. 2015 Oct 20;11(10):e1005227. doi: 10.1371/journal.ppat.1005227. eCollection 2015 Oct. PLoS Pathog. 2015. PMID: 26484870 Free PMC article.
-
Neutralizing Antibodies Limit Cell-Associated Spread of Human Cytomegalovirus in Epithelial Cells and Fibroblasts.Viruses. 2022 Jan 28;14(2):284. doi: 10.3390/v14020284. Viruses. 2022. PMID: 35215877 Free PMC article.
References
-
- Britt W (2006) Human cytomegalovirus infections and mechanisms of disease. In: Reddehase MJ, editor. Cytomegaloviruses: molecular biology and immunology. Norfolk (UK): Caister Academic Press. pp. 1–28.
-
- Kenneson A, Cannon MJ (2007) Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 17: 253–276. - PubMed
-
- Arvin AM, Fast P, Myers M, Plotkin S, Rabinovich R, et al. (2004) Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clin Infect Dis 39: 233–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

