Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data
- PMID: 25319062
- PMCID: PMC4269342
- DOI: 10.1093/annonc/mdu479
Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data
Abstract
Background: Exome or whole-genome deep sequencing of tumor DNA along with paired normal DNA can potentially provide a detailed picture of the somatic mutations that characterize the tumor. However, analysis of such sequence data can be complicated by the presence of normal cells in the tumor specimen, by intratumor heterogeneity, and by the sheer size of the raw data. In particular, determination of copy number variations from exome sequencing data alone has proven difficult; thus, single nucleotide polymorphism (SNP) arrays have often been used for this task. Recently, algorithms to estimate absolute, but not allele-specific, copy number profiles from tumor sequencing data have been described.
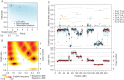
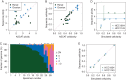
Materials and methods: We developed Sequenza, a software package that uses paired tumor-normal DNA sequencing data to estimate tumor cellularity and ploidy, and to calculate allele-specific copy number profiles and mutation profiles. We applied Sequenza, as well as two previously published algorithms, to exome sequence data from 30 tumors from The Cancer Genome Atlas. We assessed the performance of these algorithms by comparing their results with those generated using matched SNP arrays and processed by the allele-specific copy number analysis of tumors (ASCAT) algorithm.
Results: Comparison between Sequenza/exome and SNP/ASCAT revealed strong correlation in cellularity (Pearson's r = 0.90) and ploidy estimates (r = 0.42, or r = 0.94 after manual inspecting alternative solutions). This performance was noticeably superior to previously published algorithms. In addition, in artificial data simulating normal-tumor admixtures, Sequenza detected the correct ploidy in samples with tumor content as low as 30%.
Conclusions: The agreement between Sequenza and SNP array-based copy number profiles suggests that exome sequencing alone is sufficient not only for identifying small scale mutations but also for estimating cellularity and inferring DNA copy number aberrations.
Keywords: cancer genomics; copy number alterations; mutations; next-generation sequencing; software.
© The Author 2014. Published by Oxford University Press on behalf of the European Society for Medical Oncology.
Figures
Comment in
-
Pushing the boundaries of somatic copy-number variation detection: advances and challenges.Ann Oncol. 2015 Jan;26(1):11-12. doi: 10.1093/annonc/mdu536. Epub 2014 Nov 17. Ann Oncol. 2015. PMID: 25403588 No abstract available.
Similar articles
-
FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing.Nucleic Acids Res. 2016 Sep 19;44(16):e131. doi: 10.1093/nar/gkw520. Epub 2016 Jun 7. Nucleic Acids Res. 2016. PMID: 27270079 Free PMC article.
-
Ploidy- and Purity-Adjusted Allele-Specific DNA Analysis Using CLONETv2.Curr Protoc Bioinformatics. 2019 Sep;67(1):e81. doi: 10.1002/cpbi.81. Curr Protoc Bioinformatics. 2019. PMID: 31524989 Free PMC article.
-
AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data.Bioinformatics. 2014 Apr 15;30(8):1056-1063. doi: 10.1093/bioinformatics/btt759. Epub 2014 Jan 2. Bioinformatics. 2014. PMID: 24389661 Free PMC article.
-
Using SAAS-CNV to Detect and Characterize Somatic Copy Number Alterations in Cancer Genomes from Next Generation Sequencing and SNP Array Data.Methods Mol Biol. 2018;1833:29-47. doi: 10.1007/978-1-4939-8666-8_2. Methods Mol Biol. 2018. PMID: 30039361 Review.
-
Exome sequence read depth methods for identifying copy number changes.Brief Bioinform. 2015 May;16(3):380-92. doi: 10.1093/bib/bbu027. Epub 2014 Aug 28. Brief Bioinform. 2015. PMID: 25169955 Review.
Cited by
-
Frequent CHD1 deletions in prostate cancers of African American men is associated with rapid disease progression.NPJ Precis Oncol. 2024 Sep 19;8(1):208. doi: 10.1038/s41698-024-00705-8. NPJ Precis Oncol. 2024. PMID: 39294262 Free PMC article.
-
Gamma Knife Radiosurgery does not alter the copy number aberration profile in sporadic vestibular schwannoma.J Neurooncol. 2020 Sep;149(3):373-381. doi: 10.1007/s11060-020-03631-4. Epub 2020 Sep 27. J Neurooncol. 2020. PMID: 32980934 Free PMC article.
-
Tangent normalization for somatic copy-number inference in cancer genome analysis.Bioinformatics. 2022 Oct 14;38(20):4677-4686. doi: 10.1093/bioinformatics/btac586. Bioinformatics. 2022. PMID: 36040167 Free PMC article.
-
A novel Bayesian model for assessing intratumor heterogeneity of tumor infiltrating leukocytes with multi-region gene expression sequencing.bioRxiv [Preprint]. 2023 Oct 29:2023.10.24.563820. doi: 10.1101/2023.10.24.563820. bioRxiv. 2023. PMID: 37961165 Free PMC article. Preprint.
-
DiPRO1 distinctly reprograms muscle and mesenchymal cancer cells.EMBO Mol Med. 2024 Aug;16(8):1840-1885. doi: 10.1038/s44321-024-00097-z. Epub 2024 Jul 15. EMBO Mol Med. 2024. PMID: 39009887 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

