PMI: a ΔΨm independent pharmacological regulator of mitophagy
- PMID: 25455860
- PMCID: PMC4245710
- DOI: 10.1016/j.chembiol.2014.09.019
PMI: a ΔΨm independent pharmacological regulator of mitophagy
Abstract
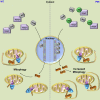
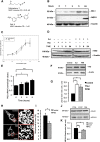
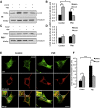
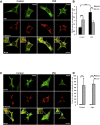
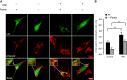
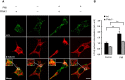
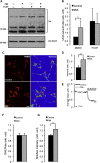
Mitophagy is central to mitochondrial and cellular homeostasis and operates via the PINK1/Parkin pathway _targeting mitochondria devoid of membrane potential (ΔΨm) to autophagosomes. Although mitophagy is recognized as a fundamental cellular process, selective pharmacologic modulators of mitophagy are almost nonexistent. We developed a compound that increases the expression and signaling of the autophagic adaptor molecule P62/SQSTM1 and forces mitochondria into autophagy. The compound, P62-mediated mitophagy inducer (PMI), activates mitophagy without recruiting Parkin or collapsing ΔΨm and retains activity in cells devoid of a fully functional PINK1/Parkin pathway. PMI drives mitochondria to a process of quality control without compromising the bio-energetic competence of the whole network while exposing just those organelles to be recycled. Thus, PMI circumvents the toxicity and some of the nonspecific effects associated with the abrupt dissipation of ΔΨm by ionophores routinely used to induce mitophagy and represents a prototype pharmacological tool to investigate the molecular mechanisms of mitophagy.
Figures
Similar articles
-
Mitochondrial and lysosomal biogenesis are activated following PINK1/parkin-mediated mitophagy.J Neurochem. 2016 Jan;136(2):388-402. doi: 10.1111/jnc.13412. Epub 2015 Nov 24. J Neurochem. 2016. PMID: 26509433 Free PMC article.
-
p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both.Autophagy. 2010 Nov;6(8):1090-106. doi: 10.4161/auto.6.8.13426. Autophagy. 2010. PMID: 20890124 Free PMC article.
-
AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1.Cell Death Differ. 2015 Mar;22(3):419-32. doi: 10.1038/cdd.2014.139. Epub 2014 Sep 12. Cell Death Differ. 2015. PMID: 25215947 Free PMC article.
-
Regulation by mitophagy.Int J Biochem Cell Biol. 2014 Aug;53:147-50. doi: 10.1016/j.biocel.2014.05.012. Epub 2014 May 16. Int J Biochem Cell Biol. 2014. PMID: 24842103 Review.
-
_targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control.Antioxid Redox Signal. 2011 May 15;14(10):1929-38. doi: 10.1089/ars.2010.3799. Epub 2011 Mar 3. Antioxid Redox Signal. 2011. PMID: 21194381 Free PMC article. Review.
Cited by
-
Mycotoxin Patulin Suppresses Innate Immune Responses by Mitochondrial Dysfunction and p62/Sequestosome-1-dependent Mitophagy.J Biol Chem. 2016 Sep 9;291(37):19299-311. doi: 10.1074/jbc.M115.686683. Epub 2016 Jul 25. J Biol Chem. 2016. PMID: 27458013 Free PMC article.
-
Insulin-Like Growth Factor I Prevents Cellular Aging via Activation of Mitophagy.J Aging Res. 2020 Aug 1;2020:4939310. doi: 10.1155/2020/4939310. eCollection 2020. J Aging Res. 2020. PMID: 32802505 Free PMC article.
-
Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation.Nutrients. 2022 Apr 19;14(9):1690. doi: 10.3390/nu14091690. Nutrients. 2022. PMID: 35565658 Free PMC article. Review.
-
Mechanisms of mitophagy in cellular homeostasis, physiology and pathology.Nat Cell Biol. 2018 Sep;20(9):1013-1022. doi: 10.1038/s41556-018-0176-2. Epub 2018 Aug 28. Nat Cell Biol. 2018. PMID: 30154567 Review.
-
MitophAging: Mitophagy in Aging and Disease.Front Cell Dev Biol. 2020 Apr 15;8:239. doi: 10.3389/fcell.2020.00239. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32373609 Free PMC article. Review.
References
-
- Abrahams J.P., Leslie A.G., Lutter R., Walker J.E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. - PubMed
-
- Baird L., Dinkova-Kostova A.T. The cytoprotective role of the Keap1-Nrf2 pathway. Arch. Toxicol. 2011;85:241–272. - PubMed
-
- Capaldi R.A. Structure and function of cytochrome c oxidase. Annu. Rev. Biochem. 1990;59:569–596. - PubMed
-
- Cheng X., Siow R.C., Mann G.E. Impaired redox signaling and antioxidant gene expression in endothelial cells in diabetes: a role for mitochondria and the nuclear factor-E2-related factor 2-Kelch-like ECH-associated protein 1 defense pathway. Antioxid. Redox Signal. 2011;14:469–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

