RIP3 induces apoptosis independent of pronecrotic kinase activity
- PMID: 25459880
- PMCID: PMC4512186
- DOI: 10.1016/j.molcel.2014.10.021
RIP3 induces apoptosis independent of pronecrotic kinase activity
Abstract
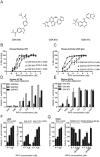
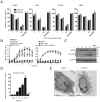
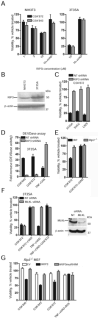
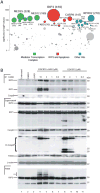
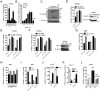
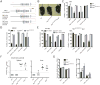
Receptor-interacting protein kinase 3 (RIP3 or RIPK3) has emerged as a central player in necroptosis and a potential _target to control inflammatory disease. Here, three selective small-molecule compounds are shown to inhibit RIP3 kinase-dependent necroptosis, although their therapeutic value is undermined by a surprising, concentration-dependent induction of apoptosis. These compounds interact with RIP3 to activate caspase 8 (Casp8) via RHIM-driven recruitment of RIP1 (RIPK1) to assemble a Casp8-FADD-cFLIP complex completely independent of pronecrotic kinase activities and MLKL. RIP3 kinase-dead D161N mutant induces spontaneous apoptosis independent of compound, whereas D161G, D143N, and K51A mutants, like wild-type, only trigger apoptosis when compound is present. Accordingly, RIP3-K51A mutant mice (Rip3(K51A/K51A)) are viable and fertile, in stark contrast to the perinatal lethality of Rip3(D161N/D161N) mice. RIP3 therefore holds both necroptosis and apoptosis in balance through a Ripoptosome-like platform. This work highlights a common mechanism unveiling RHIM-driven apoptosis by therapeutic or genetic perturbation of RIP3.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
RIP1, RIP3, and MLKL Contribute to Cell Death Caused by Clostridium perfringens Enterotoxin.mBio. 2019 Dec 17;10(6):e02985-19. doi: 10.1128/mBio.02985-19. mBio. 2019. PMID: 31848291 Free PMC article.
-
RIP3 mediates the embryonic lethality of caspase-8-deficient mice.Nature. 2011 Mar 17;471(7338):368-72. doi: 10.1038/nature09857. Epub 2011 Mar 2. Nature. 2011. PMID: 21368762 Free PMC article.
-
Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis.Science. 2014 Mar 21;343(6177):1357-60. doi: 10.1126/science.1249361. Epub 2014 Feb 20. Science. 2014. PMID: 24557836
-
Characterization of the ripoptosome and its components: implications for anti-inflammatory and cancer therapy.Methods Enzymol. 2014;545:83-102. doi: 10.1016/B978-0-12-801430-1.00004-4. Methods Enzymol. 2014. PMID: 25065887 Review.
-
Necroptosis in health and diseases.Semin Cell Dev Biol. 2014 Nov;35:14-23. doi: 10.1016/j.semcdb.2014.07.013. Epub 2014 Aug 1. Semin Cell Dev Biol. 2014. PMID: 25087983 Review.
Cited by
-
Induction and Detection of Necroptotic Cell Death in Mammalian Cell Culture.Methods Mol Biol. 2021;2255:119-134. doi: 10.1007/978-1-0716-1162-3_11. Methods Mol Biol. 2021. PMID: 34033099
-
Mediators of necroptosis: from cell death to metabolic regulation.EMBO Mol Med. 2024 Feb;16(2):219-237. doi: 10.1038/s44321-023-00011-z. Epub 2024 Jan 9. EMBO Mol Med. 2024. PMID: 38195700 Free PMC article. Review.
-
Viral dosing of influenza A infection reveals involvement of RIPK3 and FADD, but not MLKL.Cell Death Dis. 2021 May 11;12(5):471. doi: 10.1038/s41419-021-03746-0. Cell Death Dis. 2021. PMID: 33976111 Free PMC article.
-
Generation of small molecules to interfere with regulated necrosis.Cell Mol Life Sci. 2016 Jun;73(11-12):2251-67. doi: 10.1007/s00018-016-2198-x. Epub 2016 Apr 5. Cell Mol Life Sci. 2016. PMID: 27048812 Free PMC article. Review.
-
Structure-based development of potent and selective type-II kinase inhibitors of RIPK1.Acta Pharm Sin B. 2024 Jan;14(1):319-334. doi: 10.1016/j.apsb.2023.10.021. Epub 2023 Nov 8. Acta Pharm Sin B. 2024. PMID: 38261830 Free PMC article.
References
-
- Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M, Capriotti C, Cook M, Finger J, Hughes-Earle A, et al. Cutting Edge: RIP1 kinase activity is dispensable for normal development but Is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol. 2014;192:5476–5480. - PMC - PubMed
-
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. - PubMed
-
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. - PubMed
-
- Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol. 2008;9:378–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

