MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens
- PMID: 25476604
- PMCID: PMC4290824
- DOI: 10.1186/s13059-014-0554-4
MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens
Abstract
We propose the Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) method for prioritizing single-guide RNAs, genes and pathways in genome-scale CRISPR/Cas9 knockout screens. MAGeCK demonstrates better performance compared with existing methods, identifies both positively and negatively selected genes simultaneously, and reports robust results across different experimental conditions. Using public datasets, MAGeCK identified novel essential genes and pathways, including EGFR in vemurafenib-treated A375 cells harboring a BRAF mutation. MAGeCK also detected cell type-specific essential genes, including BCR and ABL1, in KBM7 cells bearing a BCR-ABL fusion, and IGF1R in HL-60 cells, which depends on the insulin signaling pathway for proliferation.
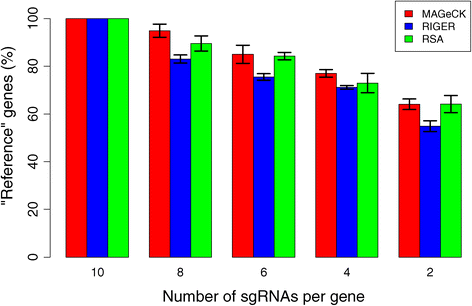
Figures
Similar articles
-
Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR.Genome Biol. 2015 Dec 16;16:281. doi: 10.1186/s13059-015-0843-6. Genome Biol. 2015. PMID: 26673418 Free PMC article.
-
Unsupervised correction of gene-independent cell responses to CRISPR-Cas9 _targeting.BMC Genomics. 2018 Aug 13;19(1):604. doi: 10.1186/s12864-018-4989-y. BMC Genomics. 2018. PMID: 30103702 Free PMC article.
-
CRISPR-Cas9 screens reveal common essential miRNAs in human cancer cell lines.Genome Med. 2024 Jun 17;16(1):82. doi: 10.1186/s13073-024-01341-4. Genome Med. 2024. PMID: 38886809 Free PMC article.
-
Functional genomic screening approaches in mechanistic toxicology and potential future applications of CRISPR-Cas9.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:31-42. doi: 10.1016/j.mrrev.2015.01.002. Epub 2015 Jan 25. Mutat Res Rev Mutat Res. 2015. PMID: 26041264 Free PMC article. Review.
-
Development and potential applications of CRISPR-Cas9 genome editing technology in sarcoma.Cancer Lett. 2016 Apr 1;373(1):109-118. doi: 10.1016/j.canlet.2016.01.030. Epub 2016 Jan 21. Cancer Lett. 2016. PMID: 26806808 Free PMC article. Review.
Cited by
-
NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism.Nat Metab. 2020 Apr;2(4):318-334. doi: 10.1038/s42255-020-0191-z. Epub 2020 Apr 20. Nat Metab. 2020. PMID: 32691018 Free PMC article.
-
Brca2 deficiency drives gastrointestinal tumor formation and is selectively inhibited by mitomycin C.Cell Death Dis. 2020 Sep 26;11(9):812. doi: 10.1038/s41419-020-03013-8. Cell Death Dis. 2020. PMID: 32980867 Free PMC article.
-
RNA Binding Proteins As Regulators of Oxidative Stress Identified by a _targeted CRISPR-Cas9 Single Guide RNA Library.CRISPR J. 2021 Jun;4(3):427-437. doi: 10.1089/crispr.2020.0116. Epub 2021 Jun 4. CRISPR J. 2021. PMID: 34096786 Free PMC article.
-
A Dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus.Sci Rep. 2015 Mar 9;5:8865. doi: 10.1038/srep08865. Sci Rep. 2015. PMID: 25746010 Free PMC article.
-
A genome-wide CRISPR screen identifies UFMylation and TRAMP-like complexes as host factors required for hepatitis A virus infection.Cell Rep. 2021 Mar 16;34(11):108859. doi: 10.1016/j.celrep.2021.108859. Cell Rep. 2021. PMID: 33730579 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

