A host susceptibility gene, DR1, facilitates influenza A virus replication by suppressing host innate immunity and enhancing viral RNA replication
- PMID: 25589657
- PMCID: PMC4403386
- DOI: 10.1128/JVI.03610-14
A host susceptibility gene, DR1, facilitates influenza A virus replication by suppressing host innate immunity and enhancing viral RNA replication
Abstract
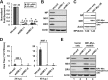
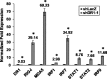
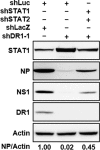
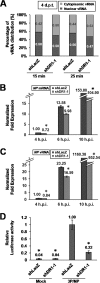
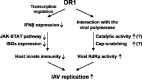
Influenza A virus (IAV) depends on cellular factors to complete its replication cycle; thus, investigation of the factors utilized by IAV may facilitate antiviral drug development. To this end, a cellular transcriptional repressor, DR1, was identified from a genome-wide RNA interference (RNAi) screen. Knockdown (KD) of DR1 resulted in reductions of viral RNA and protein production, demonstrating that DR1 acts as a positive host factor in IAV replication. Genome-wide transcriptomic analysis showed that there was a strong induction of interferon-stimulated gene (ISG) expression after prolonged DR1 KD. We found that beta interferon (IFN-β) was induced by DR1 KD, thereby activating the JAK-STAT pathway to turn on ISG expression, which led to a strong inhibition of IAV replication. This result suggests that DR1 in normal cells suppresses IFN induction, probably to prevent undesired cytokine production, but that this suppression may create a milieu that favors IAV replication once cells are infected. Furthermore, biochemical assays of viral RNA replication showed that DR1 KD suppressed viral RNA replication. We also showed that DR1 associated with all three subunits of the viral RNA-dependent RNA polymerase (RdRp) complex, indicating that DR1 may interact with individual components of the viral RdRp complex to enhance viral RNA replication. Thus, DR1 may be considered a novel host susceptibility gene for IAV replication via a dual mechanism, not only suppressing the host defense to indirectly favor IAV replication but also directly facilitating viral RNA replication.
Importance: Investigations of virus-host interactions involved in influenza A virus (IAV) replication are important for understanding viral pathogenesis and host defenses, which may manipulate influenza virus infection or prevent the emergence of drug resistance caused by a high error rate during viral RNA replication. For this purpose, a cellular transcriptional repressor, DR1, was identified from a genome-wide RNAi screen as a positive regulator in IAV replication. In the current studies, we showed that DR1 suppressed the gene expression of a large set of host innate immunity genes, which indirectly facilitated IAV replication in the event of IAV infection. Besides this scenario, DR1 also directly enhanced the viral RdRp activity, likely through associating with individual components of the viral RdRp complex. Thus, DR1 represents a novel host susceptibility gene for IAV replication via multiple functions, not only suppressing the host defense but also enhancing viral RNA replication. DR1 may be a potential _target for drug development against influenza virus infection.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Zinc Finger-Containing Cellular Transcription Corepressor ZBTB25 Promotes Influenza Virus RNA Transcription and Is a _target for Zinc Ejector Drugs.J Virol. 2017 Sep 27;91(20):e00842-17. doi: 10.1128/JVI.00842-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28768860 Free PMC article.
-
Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression.J Virol. 2014 Aug;88(15):8375-85. doi: 10.1128/JVI.00126-14. Epub 2014 May 14. J Virol. 2014. PMID: 24829357 Free PMC article.
-
Cellular 5'-3' mRNA Exoribonuclease XRN1 Inhibits Interferon Beta Activation and Facilitates Influenza A Virus Replication.mBio. 2021 Aug 31;12(4):e0094521. doi: 10.1128/mBio.00945-21. Epub 2021 Jul 27. mBio. 2021. PMID: 34311580 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Induction of innate immunity and its perturbation by influenza viruses.Protein Cell. 2015 Oct;6(10):712-21. doi: 10.1007/s13238-015-0191-z. Epub 2015 Jul 24. Protein Cell. 2015. PMID: 26206138 Free PMC article. Review.
Cited by
-
Upregulation of galectin-3 in influenza A virus infection promotes viral RNA synthesis through its association with viral PA protein.J Biomed Sci. 2023 Feb 23;30(1):14. doi: 10.1186/s12929-023-00901-x. J Biomed Sci. 2023. PMID: 36823664 Free PMC article.
-
RSAD2 Is an Effective _target for High-Yield Vaccine Production in MDCK Cells.Viruses. 2022 Nov 21;14(11):2587. doi: 10.3390/v14112587. Viruses. 2022. PMID: 36423196 Free PMC article.
-
Up-regulation of microRNA-203 in influenza A virus infection inhibits viral replication by _targeting DR1.Sci Rep. 2018 May 1;8(1):6797. doi: 10.1038/s41598-018-25073-9. Sci Rep. 2018. PMID: 29717211 Free PMC article.
-
To Conquer the Host, Influenza Virus Is Packing It In: Interferon-Antagonistic Strategies beyond NS1.J Virol. 2016 Sep 12;90(19):8389-94. doi: 10.1128/JVI.00041-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27440898 Free PMC article. Review.
-
Tripartite motif-containing protein 46 accelerates influenza A H7N9 virus infection by promoting K48-linked ubiquitination of TBK1.Virol J. 2022 Nov 3;19(1):176. doi: 10.1186/s12985-022-01907-x. Virol J. 2022. PMID: 36329446 Free PMC article.
References
-
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi:10.1128/JVI.79.5.2814-2822.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

