A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation
- PMID: 25737281
- PMCID: PMC4358404
- DOI: 10.1101/gad.254532.114
A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation
Abstract
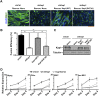
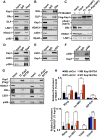
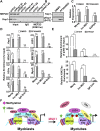
The transcriptional activator MyoD serves as a master controller of myogenesis. Often in partnership with Mef2 (myocyte enhancer factor 2), MyoD binds to the promoters of hundreds of muscle genes in proliferating myoblasts yet activates these _targets only upon receiving cues that launch differentiation. What regulates this off/on switch of MyoD function has been incompletely understood, although it is known to reflect the action of chromatin modifiers. Here, we identify KAP1 (KRAB [Krüppel-like associated box]-associated protein 1)/TRIM28 (tripartite motif protein 28) as a key regulator of MyoD function. In myoblasts, KAP1 is present with MyoD and Mef2 at many muscle genes, where it acts as a scaffold to recruit not only coactivators such as p300 and LSD1 but also corepressors such as G9a and HDAC1 (histone deacetylase 1), with promoter silencing as the net outcome. Upon differentiation, MSK1-mediated phosphorylation of KAP1 releases the corepressors from the scaffold, unleashing transcriptional activation by MyoD/Mef2 and their positive cofactors. Thus, our results reveal KAP1 as a previously unappreciated interpreter of cell signaling, which modulates the ability of MyoD to drive myogenesis.
Keywords: G9a; KAP1; MSK1 phosphorylation; MyoD; epigenetics; myogenesis.
© 2015 Singh et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Similar articles
-
Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis.Mol Cell. 2001 Oct;8(4):885-97. doi: 10.1016/s1097-2765(01)00373-2. Mol Cell. 2001. PMID: 11684023
-
PC4 coactivates MyoD by relieving the histone deacetylase 4-mediated inhibition of myocyte enhancer factor 2C.Mol Cell Biol. 2005 Mar;25(6):2242-59. doi: 10.1128/MCB.25.6.2242-2259.2005. Mol Cell Biol. 2005. PMID: 15743821 Free PMC article.
-
The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis.Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7354-9. doi: 10.1073/pnas.131198498. Epub 2001 Jun 5. Proc Natl Acad Sci U S A. 2001. PMID: 11390982 Free PMC article.
-
Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle.Epigenetics. 2010 Nov-Dec;5(8):691-5. doi: 10.4161/epi.5.8.13045. Epub 2010 Nov 1. Epigenetics. 2010. PMID: 20716948 Free PMC article. Review.
-
MEF2: a transcriptional _target for signaling pathways controlling skeletal muscle growth and differentiation.Curr Opin Cell Biol. 1999 Dec;11(6):683-8. doi: 10.1016/s0955-0674(99)00036-8. Curr Opin Cell Biol. 1999. PMID: 10600704 Review.
Cited by
-
Canonical Wnt signalling regulates nuclear export of Setdb1 during skeletal muscle terminal differentiation.Cell Discov. 2016 Oct 18;2:16037. doi: 10.1038/celldisc.2016.37. eCollection 2016. Cell Discov. 2016. PMID: 27790377 Free PMC article.
-
KAP1 phosphorylation promotes the survival of neural stem cells after ischemia/reperfusion by maintaining the stability of PCNA.Stem Cell Res Ther. 2022 Jul 7;13(1):290. doi: 10.1186/s13287-022-02962-5. Stem Cell Res Ther. 2022. PMID: 35799276 Free PMC article.
-
Phosphorylation of TRIM28 Enhances the Expression of IFN-β and Proinflammatory Cytokines During HPAIV Infection of Human Lung Epithelial Cells.Front Immunol. 2018 Sep 28;9:2229. doi: 10.3389/fimmu.2018.02229. eCollection 2018. Front Immunol. 2018. PMID: 30323812 Free PMC article.
-
Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues.Dev Cell. 2016 Mar 21;36(6):611-23. doi: 10.1016/j.devcel.2016.02.024. Dev Cell. 2016. PMID: 27003935 Free PMC article.
-
Deletion of Trim28 in committed adipocytes promotes obesity but preserves glucose tolerance.Nat Commun. 2021 Jan 4;12(1):74. doi: 10.1038/s41467-020-20434-3. Nat Commun. 2021. PMID: 33397965 Free PMC article.
References
-
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. 2008. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation (N-ChIP) of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc 3: 398–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
