Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1
- PMID: 25759303
- PMCID: PMC4615252
- DOI: 10.1080/19491034.2014.1003512
Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1
Abstract
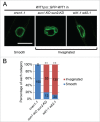
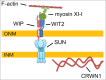
Nuclei undergo dynamic shape changes during plant development, but the mechanism is unclear. In Arabidopsis, Sad1/UNC-84 (SUN) proteins, WPP domain-interacting proteins (WIPs), WPP domain-interacting tail-anchored proteins (WITs), myosin XI-i, and CROWDED NUCLEI 1 (CRWN1) have been shown to be essential for nuclear elongation in various epidermal cell types. It has been proposed that WITs serve as adaptors linking myosin XI-i to the SUN-WIP complex at the nuclear envelope (NE). Recently, an interaction between Arabidopsis SUN1 and SUN2 proteins and CRWN1, a plant analog of lamins, has been reported. Therefore, the CRWN1-SUN-WIP-WIT-myosin XI-i interaction may form a linker of the nucleoskeleton to the cytoskeleton complex. In this study, we investigate this proposed mechanism in detail for nuclei of Arabidopsis root hairs and trichomes. We show that WIT2, but not WIT1, plays an essential role in nuclear shape determination by recruiting myosin XI-i to the SUN-WIP NE bridges. Compared with SUN2, SUN1 plays a predominant role in nuclear shape. The NE localization of SUN1, SUN2, WIP1, and a truncated WIT2 does not depend on CRWN1. While crwn1 mutant nuclei are smooth, the nuclei of sun or wit mutants are invaginated, similar to the reported myosin XI-i mutant phenotype. Together, this indicates that the roles of the respective WIT and SUN paralogs have diverged in trichomes and root hairs, and that the SUN-WIP-WIT2-myosin XI-i complex and CRWN1 independently determine elongated nuclear shape. This supports a model of nuclei being shaped both by cytoplasmic forces transferred to the NE and by nucleoplasmic filaments formed under the NE.
Keywords: Arabidopsis; CDS, coding sequence; CRWN; CRWN1, CROWDED NUCLEI 1; KASH; KASH, Klarsicht/ANC-1/Syne-1 Homology; LINC; LINC, linker of the nucleoskeleton to the cytoskeleton; NE, nuclear envelope; NLI, nuclear envelope localization index; SUN; SUN, Sad1/UNC-84; WIP, WPP domain-interacting protein; WIT, WPP domain-interacting tail-anchored protein; XI-iC642, myosin XI-i C-terminal 642 amino acids.; nuclear envelope; nuclear shape; sun1-KO sun2-KD, sun1-knockout sun2-knockdown.
Figures
Similar articles
-
SUN anchors pollen WIP-WIT complexes at the vegetative nuclear envelope and is necessary for pollen tube _targeting and fertility.J Exp Bot. 2015 Dec;66(22):7299-307. doi: 10.1093/jxb/erv425. Epub 2015 Sep 25. J Exp Bot. 2015. PMID: 26409047 Free PMC article.
-
Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination.J Cell Biol. 2012 Jan 23;196(2):203-11. doi: 10.1083/jcb.201108098. J Cell Biol. 2012. PMID: 22270916 Free PMC article.
-
The plant LINC complex at the nuclear envelope.Chromosome Res. 2014 Jun;22(2):241-52. doi: 10.1007/s10577-014-9419-7. Chromosome Res. 2014. PMID: 24801343 Review.
-
Integrity of the Linker of Nucleoskeleton and Cytoskeleton Is Required for Efficient Herpesvirus Nuclear Egress.J Virol. 2017 Sep 12;91(19):e00330-17. doi: 10.1128/JVI.00330-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28724767 Free PMC article.
-
Protein interactions at the higher plant nuclear envelope: evidence for a linker of nucleoskeleton and cytoskeleton complex.Front Plant Sci. 2014 May 7;5:183. doi: 10.3389/fpls.2014.00183. eCollection 2014. Front Plant Sci. 2014. PMID: 24847341 Free PMC article. Review.
Cited by
-
Exploring the evolution of the proteins of the plant nuclear envelope.Nucleus. 2017 Jan 2;8(1):46-59. doi: 10.1080/19491034.2016.1236166. Epub 2016 Sep 19. Nucleus. 2017. PMID: 27644504 Free PMC article.
-
Nuclear migration events throughout development.J Cell Sci. 2016 May 15;129(10):1951-61. doi: 10.1242/jcs.179788. J Cell Sci. 2016. PMID: 27182060 Free PMC article. Review.
-
Plant nuclear envelope as a hub connecting genome organization with regulation of gene expression.Nucleus. 2023 Dec;14(1):2178201. doi: 10.1080/19491034.2023.2178201. Nucleus. 2023. PMID: 36794966 Free PMC article.
-
SUN anchors pollen WIP-WIT complexes at the vegetative nuclear envelope and is necessary for pollen tube _targeting and fertility.J Exp Bot. 2015 Dec;66(22):7299-307. doi: 10.1093/jxb/erv425. Epub 2015 Sep 25. J Exp Bot. 2015. PMID: 26409047 Free PMC article.
-
Distinct Roles for KASH Proteins SINE1 and SINE2 in Guard Cell Actin Reorganization, Calcium Oscillations, and Vacuolar Remodeling.Front Plant Sci. 2022 May 6;13:784342. doi: 10.3389/fpls.2022.784342. eCollection 2022. Front Plant Sci. 2022. PMID: 35599883 Free PMC article.
References
-
- Gladfelter A, Berman J. Dancing genomes: fungal nuclear positioning. Nat Rev Microbiol 2009; 7:875-86; PMID:19898490; http://dx.doi.org/10.1038/nrmicro2249 - DOI - PMC - PubMed
-
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 2010; 26:421-44; PMID:20507227; http://dx.doi.org/10.1146/annurev-cellbio-100109-104037 - DOI - PMC - PubMed
-
- Gundersen GG, Worman HJ. Nuclear positioning. Cell 2013; 152:1376-89; PMID:23498944; http://dx.doi.org/10.1016/j.cell.2013.02.031 - DOI - PMC - PubMed
-
- Mejat A, Misteli T. LINC complexes in health and disease. Nucleus 2010; 1:40-52; PMID:21327104; http://dx.doi.org/10.4161/nucl.1.1.10530 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
