Non-coding RNA: a new frontier in regulatory biology
- PMID: 25821635
- PMCID: PMC4374487
- DOI: 10.1093/nsr/nwu008
Non-coding RNA: a new frontier in regulatory biology
Abstract
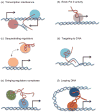
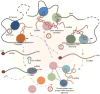
A striking finding in the past decade is the production of numerous non-coding RNAs (ncRNAs) from mammalian genomes. While it is entirely possible that many of those ncRNAs are transcription noises or by-products of RNA processing, increasing evidence suggests that a large fraction of them are functional and provide various regulatory activities in the cell. Thus, functional genomics and proteomics are incomplete without understanding functional ribonomics. As has been long suggested by the 'RNA world' hypothesis, many ncRNAs have the capacity to act like proteins in diverse biochemical processes. The enormous amount of information residing in the primary sequences and secondary structures of ncRNAs makes them particularly suited to function as scaffolds for molecular interactions. In addition, their functions appear to be stringently controlled by default via abundant nucleases when not engaged in specific interactions. This review focuses on the functional properties of regulatory ncRNAs in comparison with proteins and emphasizes both the opportunities and challenges in future ncRNA research.
Keywords: biological functions; experimental approaches; functional genomics; non-coding RNA; regulatory mechanisms; the RNA world.
Figures

Similar articles
-
Long Non-coding RNA in Plants in the Era of Reference Sequences.Front Plant Sci. 2020 Mar 12;11:276. doi: 10.3389/fpls.2020.00276. eCollection 2020. Front Plant Sci. 2020. PMID: 32226437 Free PMC article. Review.
-
[Non-coding RNA in fungi--a review].Wei Sheng Wu Xue Bao. 2013 Aug 4;53(8):790-7. Wei Sheng Wu Xue Bao. 2013. PMID: 24341270 Review. Chinese.
-
Encoding activities of non-coding RNAs.Theranostics. 2018 Mar 28;8(9):2496-2507. doi: 10.7150/thno.24677. eCollection 2018. Theranostics. 2018. PMID: 29721095 Free PMC article. Review.
-
Molecular Functions of Long Non-Coding RNAs in Plants.Genes (Basel). 2012 Mar 8;3(1):176-90. doi: 10.3390/genes3010176. Genes (Basel). 2012. PMID: 24704849 Free PMC article.
-
Genomic features and regulatory roles of intermediate-sized non-coding RNAs in Arabidopsis.Mol Plant. 2014 Mar;7(3):514-27. doi: 10.1093/mp/sst177. Epub 2014 Jan 7. Mol Plant. 2014. PMID: 24398630
Cited by
-
Genome and Comparative Transcriptome Dissection Provide Insights Into Molecular Mechanisms of Sclerotium Formation in Culinary-Medicinal Mushroom Pleurotus tuber-regium.Front Microbiol. 2022 Feb 17;12:815954. doi: 10.3389/fmicb.2021.815954. eCollection 2021. Front Microbiol. 2022. PMID: 35250915 Free PMC article.
-
Long Noncoding RNAs: New Players in the Osteogenic Differentiation of Bone Marrow- and Adipose-Derived Mesenchymal Stem Cells.Stem Cell Rev Rep. 2018 Jun;14(3):297-308. doi: 10.1007/s12015-018-9801-5. Stem Cell Rev Rep. 2018. PMID: 29464508 Review.
-
Preliminary RNA-Seq Analysis of Long Non-Coding RNAs Expressed in Human Term Placenta.Int J Mol Sci. 2018 Jun 27;19(7):1894. doi: 10.3390/ijms19071894. Int J Mol Sci. 2018. PMID: 29954144 Free PMC article.
-
Long non-coding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b.Protein Cell. 2020 Mar;11(3):161-186. doi: 10.1007/s13238-019-0650-z. Epub 2019 Jul 17. Protein Cell. 2020. PMID: 31317506 Free PMC article.
-
Novel role of long non-coding RNAs in autoimmune cutaneous disease.J Cell Commun Signal. 2022 Dec;16(4):487-504. doi: 10.1007/s12079-021-00639-x. Epub 2021 Aug 3. J Cell Commun Signal. 2022. PMID: 34346026 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
