Diversification of Paralogous α-Isopropylmalate Synthases by Modulation of Feedback Control and Hetero-Oligomerization in Saccharomyces cerevisiae
- PMID: 25841022
- PMCID: PMC4452578
- DOI: 10.1128/EC.00033-15
Diversification of Paralogous α-Isopropylmalate Synthases by Modulation of Feedback Control and Hetero-Oligomerization in Saccharomyces cerevisiae
Abstract
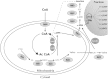
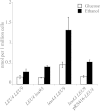
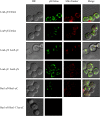
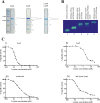
Production of α-isopropylmalate (α-IPM) is critical for leucine biosynthesis and for the global control of metabolism. The budding yeast Saccharomyces cerevisiae has two paralogous genes, LEU4 and LEU9, that encode α-IPM synthase (α-IPMS) isozymes. Little is known about the biochemical differences between these two α-IPMS isoenzymes. Here, we show that the Leu4 homodimer is a leucine-sensitive isoform, while the Leu9 homodimer is resistant to such feedback inhibition. The leu4Δ mutant, which expresses only the feedback-resistant Leu9 homodimer, grows slowly with either glucose or ethanol and accumulates elevated pools of leucine; this phenotype is alleviated by the addition of leucine. Transformation of the leu4Δ mutant with a centromeric plasmid carrying LEU4 restored the wild-type phenotype. Bimolecular fluorescent complementation analysis showed that Leu4-Leu9 heterodimeric isozymes are formed in vivo. Purification and kinetic analysis showed that the hetero-oligomeric isozyme has a distinct leucine sensitivity behavior. Determination of α-IPMS activity in ethanol-grown cultures showed that α-IPM biosynthesis and growth under these respiratory conditions depend on the feedback-sensitive Leu4 homodimer. We conclude that retention and further diversification of two yeast α-IPMSs have resulted in a specific regulatory system that controls the leucine-α-IPM biosynthetic pathway by selective feedback sensitivity of homomeric and heterodimeric isoforms.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Identification by functional analysis of the gene encoding alpha-isopropylmalate synthase II (LEU9) in Saccharomyces cerevisiae.Yeast. 2000 Apr;16(6):539-45. doi: 10.1002/(SICI)1097-0061(200004)16:6<539::AID-YEA547>3.0.CO;2-K. Yeast. 2000. PMID: 10790691
-
Influence of mutation in the regulatory domain of α-isopropylmalate synthase from Saccharomyces cerevisiae on its activity and feedback inhibition.Biosci Biotechnol Biochem. 2022 May 24;86(6):755-762. doi: 10.1093/bbb/zbac045. Biosci Biotechnol Biochem. 2022. PMID: 35333283
-
Isolation and characterization of awamori yeast mutants with L-leucine accumulation that overproduce isoamyl alcohol.J Biosci Bioeng. 2015 Feb;119(2):140-7. doi: 10.1016/j.jbiosc.2014.06.020. Epub 2014 Jul 21. J Biosci Bioeng. 2015. PMID: 25060730
-
Two alpha isopropylmalate synthase isozymes with similar kinetic properties are extant in the yeast Lachancea kluyveri.FEMS Yeast Res. 2022 Apr 8;22(1):foac016. doi: 10.1093/femsyr/foac016. FEMS Yeast Res. 2022. PMID: 35266531
-
Leucine biosynthesis in fungi: entering metabolism through the back door.Microbiol Mol Biol Rev. 2003 Mar;67(1):1-15, table of contents. doi: 10.1128/MMBR.67.1.1-15.2003. Microbiol Mol Biol Rev. 2003. PMID: 12626680 Free PMC article. Review.
Cited by
-
Molecular _targets for antifungals in amino acid and protein biosynthetic pathways.Amino Acids. 2021 Jul;53(7):961-991. doi: 10.1007/s00726-021-03007-6. Epub 2021 Jun 3. Amino Acids. 2021. PMID: 34081205 Free PMC article. Review.
-
Neo-functionalization in Saccharomyces cerevisiae: a novel Nrg1-Rtg3 chimeric transcriptional modulator is essential to maintain mitochondrial DNA integrity.R Soc Open Sci. 2023 Nov 1;10(11):231209. doi: 10.1098/rsos.231209. eCollection 2023 Nov. R Soc Open Sci. 2023. PMID: 37920568 Free PMC article.
-
Leucine biosynthesis is required for infection-related morphogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae.Curr Genet. 2020 Feb;66(1):155-171. doi: 10.1007/s00294-019-01009-2. Epub 2019 Jul 1. Curr Genet. 2020. PMID: 31263943
-
Bacilli glutamate dehydrogenases diverged via coevolution of transcription and enzyme regulation.EMBO Rep. 2017 Jul;18(7):1139-1149. doi: 10.15252/embr.201743990. Epub 2017 May 3. EMBO Rep. 2017. PMID: 28468957 Free PMC article.
-
Regulation of the Leucine Metabolism in Mortierella alpina.J Fungi (Basel). 2022 Feb 18;8(2):196. doi: 10.3390/jof8020196. J Fungi (Basel). 2022. PMID: 35205950 Free PMC article.
References
-
- Beltzer JP, Morris SR, Kohlhaw GB. 1988. Yeast LEU4 encodes mitochondrial and nonmitochondrial forms of alpha-isopropylmalate synthase. J Biol Chem 263:368–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

