Comparative analysis of the silk gland transcriptomes between the domestic and wild silkworms
- PMID: 25887670
- PMCID: PMC4328555
- DOI: 10.1186/s12864-015-1287-9
Comparative analysis of the silk gland transcriptomes between the domestic and wild silkworms
Abstract
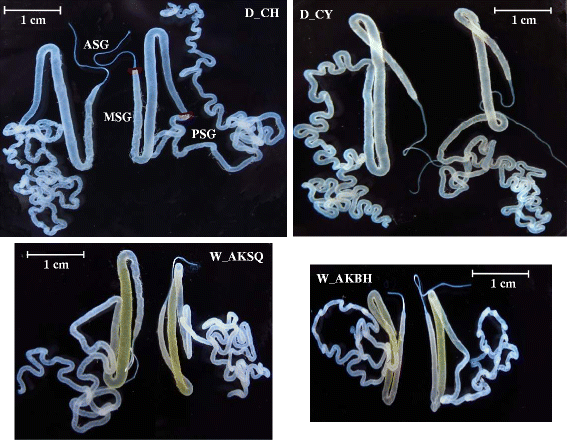
Background: Bombyx mori was domesticated from the Chinese wild silkworm, Bombyx mandarina. Wild and domestic silkworms are good models in which to investigate genes related to silk protein synthesis that may be differentially expressed in silk glands, because their silk productions are very different. Here we used the mRNA deep sequencing (RNA-seq) approach to identify the differentially expressed genes (DEGs) in the transcriptomes of the median/posterior silk glands of two domestic and two wild silkworms.
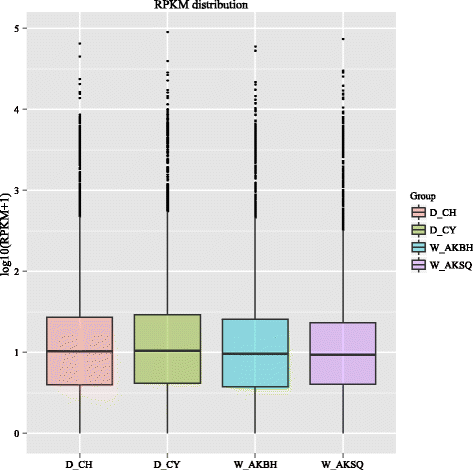
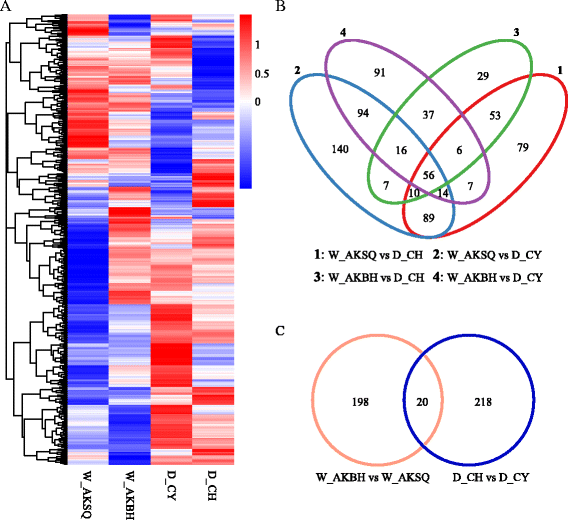
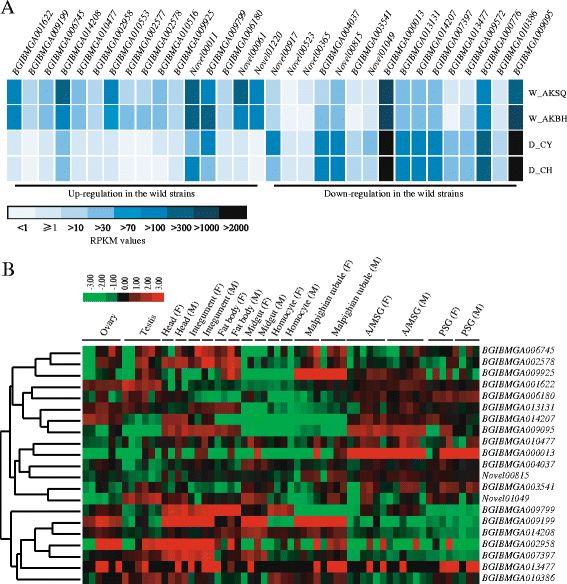
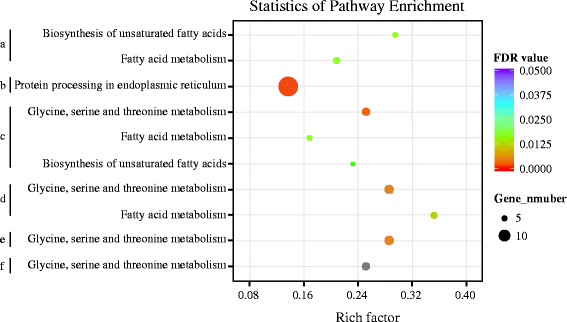
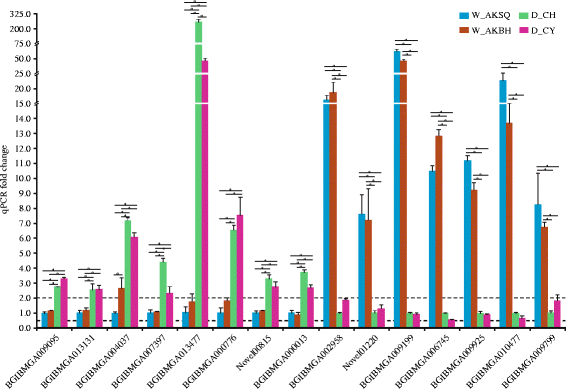
Results: The results indicated that about 58% of the total genes were expressed (reads per kilo bases per million reads (RPKM) ≥ 1) in each silkworm. Comparisons of the domestic and wild silkworm transcriptomes revealed 32 DEGs, of which 16 were up-regulated in the domestic silkworms compared with in the wild silkworms, and the other 16 were up-regulated in the wild silkworms compared with in the domestic silkworms. Quantitative real-time polymerase chain reaction (qPCR) was performed for 15 randomly selected DEGs in domestic versus wild silkworms. The qPCR results were mostly consistent with the expression levels determined from the RNA-seq data. Based on a Gene Ontology (GO) enrichment analysis and manual annotation, five of the up-regulated DEGs in the wild silkworms were predicted to be involved in immune response, and seven of the up-regulated DEGs were related to the GO term "oxidoreductase activity", which is associated with antioxidant systems. In the domestic silkworms, the up-regulated DEGs were related mainly to tissue development, secretion of proteins and metabolism.
Conclusions: The up-regulated DEGs in the two domestic silkworms may be involved mainly in the highly efficient biosynthesis and secretion of silk proteins, while the up-regulated DEGs in the two wild silkworms may play more important roles in tolerance to pathogens and environment adaptation. Our results provide a foundation for understanding the molecular mechanisms of the silk production difference between domestic and wild silkworms.
Figures
Similar articles
-
Identification and comparison of long non-coding RNAs in the silk gland between domestic and wild silkworms.Insect Sci. 2018 Aug;25(4):604-616. doi: 10.1111/1744-7917.12443. Epub 2017 May 16. Insect Sci. 2018. PMID: 28111905
-
Comparative Proteomic Analysis of Posterior Silk Glands of Wild and Domesticated Silkworms Reveals Functional Evolution during Domestication.J Proteome Res. 2017 Jul 7;16(7):2495-2507. doi: 10.1021/acs.jproteome.7b00077. Epub 2017 Jun 12. J Proteome Res. 2017. PMID: 28569067
-
Comparative Transcriptome Analysis Reveals Different Silk Yields of Two Silkworm Strains.PLoS One. 2016 May 9;11(5):e0155329. doi: 10.1371/journal.pone.0155329. eCollection 2016. PLoS One. 2016. PMID: 27159277 Free PMC article.
-
Transgenic silkworms that weave recombinant proteins into silk cocoons.Biotechnol Lett. 2011 Apr;33(4):645-54. doi: 10.1007/s10529-010-0498-z. Epub 2010 Dec 24. Biotechnol Lett. 2011. PMID: 21184136 Review.
-
The advances and perspectives of recombinant protein production in the silk gland of silkworm Bombyx mori.Transgenic Res. 2014 Oct;23(5):697-706. doi: 10.1007/s11248-014-9826-8. Epub 2014 Aug 12. Transgenic Res. 2014. PMID: 25113390 Review.
Cited by
-
Molecular Characterization of the Functional Genes Associated with Silk Assembly, Transport, and Protection in the Silk Glands of Popular Multivoltine Breeds of Silkworm Bombyx mori. L.Appl Biochem Biotechnol. 2023 Apr;195(4):2371-2394. doi: 10.1007/s12010-022-04158-2. Epub 2022 Sep 23. Appl Biochem Biotechnol. 2023. PMID: 36149583
-
Transcriptome response to elevated atmospheric CO2 concentration in the Formosan subterranean termite, Coptotermes formosanus Shiraki (Isoptera: Rhinotermitidae).PeerJ. 2016 Oct 4;4:e2527. doi: 10.7717/peerj.2527. eCollection 2016. PeerJ. 2016. PMID: 27761326 Free PMC article.
-
Decrease of gene expression diversity during domestication of animals and plants.BMC Evol Biol. 2019 Jan 11;19(1):19. doi: 10.1186/s12862-018-1340-9. BMC Evol Biol. 2019. PMID: 30634914 Free PMC article.
-
Comparative transcriptome provides insights into the selection adaptation between wild and farmed foxes.Ecol Evol. 2021 Aug 30;11(19):13475-13486. doi: 10.1002/ece3.8071. eCollection 2021 Oct. Ecol Evol. 2021. PMID: 34646484 Free PMC article.
-
Evidence of peripheral olfactory impairment in the domestic silkworms: insight from the comparative transcriptome and population genetics.BMC Genomics. 2018 Nov 1;19(1):788. doi: 10.1186/s12864-018-5172-1. BMC Genomics. 2018. PMID: 30382813 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

