Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme
- PMID: 25926653
- PMCID: PMC4473545
- DOI: 10.1128/JVI.00854-15
Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme
Abstract
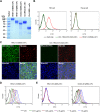
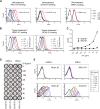
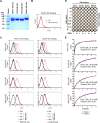
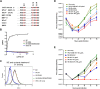
Human coronavirus (hCoV) HKU1 is one of six hCoVs identified to date and the only one with an unidentified cellular receptor. hCoV-HKU1 encodes a hemagglutinin-esterase (HE) protein that is unique to the group a betacoronaviruses (group 2a). The function of HKU1-HE remains largely undetermined. In this study, we examined binding of the S1 domain of hCoV-HKU1 spike to a panel of cells and found that the S1 could specifically bind on the cell surface of a human rhabdomyosarcoma cell line, RD. Pretreatment of RD cells with neuraminidase (NA) and trypsin greatly reduced the binding, suggesting that the binding was mediated by sialic acids on glycoproteins. However, unlike other group 2a CoVs, e.g., hCoV-OC43, for which 9-O-acetylated sialic acid (9-O-Ac-Sia) serves as a receptor determinant, HKU1-S1 bound with neither 9-O-Ac-Sia-containing glycoprotein(s) nor rat and mouse erythrocytes. Nonetheless, the HKU1-HE was similar to OC43-HE, also possessed sialate-O-acetylesterase activity, and acted as a receptor-destroying enzyme (RDE) capable of eliminating the binding of HKU1-S1 to RD cells, whereas the O-acetylesterase-inactive HKU1-HE mutant lost this capacity. Using primary human ciliated airway epithelial (HAE) cell cultures, the only in vitro replication model for hCoV-HKU1 infection, we confirmed that pretreatment of HAE cells with HE but not the enzymatically inactive mutant blocked hCoV-HKU1 infection. These results demonstrate that hCoV-HKU1 exploits O-Ac-Sia as a cellular attachment receptor determinant to initiate the infection of host cells and that its HE protein possesses the corresponding sialate-O-acetylesterase RDE activity.
Importance: Human coronaviruses (hCoV) are important human respiratory pathogens. Among the six hCoVs identified to date, only hCoV-HKU1 has no defined cellular receptor. It is also unclear whether hemagglutinin-esterase (HE) protein plays a role in viral entry. In this study, we found that, similarly to other members of the group 2a CoVs, sialic acid moieties on glycoproteins are critical receptor determinants for the hCoV-HKU1 infection. Interestingly, the virus seems to employ a type of sialic acid different from those employed by other group 2a CoVs. In addition, we determined that the HKU1-HE protein is an O-acetylesterase and acts as a receptor-destroying enzyme (RDE) for hCoV-HKU1. This is the first study to demonstrate that hCoV-HKU1 uses certain types of O-acetylated sialic acid residues on glycoproteins to initiate the infection of host cells and that the HKU1-HE protein possesses sialate-O-acetylesterase RDE activity.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.Proc Natl Acad Sci U S A. 2019 Feb 12;116(7):2681-2690. doi: 10.1073/pnas.1809667116. Epub 2019 Jan 24. Proc Natl Acad Sci U S A. 2019. PMID: 30679277 Free PMC article.
-
Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25759-25770. doi: 10.1073/pnas.2006299117. Epub 2020 Sep 29. Proc Natl Acad Sci U S A. 2020. PMID: 32994342 Free PMC article.
-
Identification of the Receptor-Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1.J Virol. 2015 Sep;89(17):8816-27. doi: 10.1128/JVI.03737-14. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085157 Free PMC article.
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
-
Roles of Sialyl Glycans in HCoV-OC43, HCoV-HKU1, MERS-CoV and SARS-CoV-2 Infections.Methods Mol Biol. 2022;2556:243-271. doi: 10.1007/978-1-0716-2635-1_17. Methods Mol Biol. 2022. PMID: 36175638 Review.
Cited by
-
Update on Human Rhinovirus and Coronavirus Infections.Semin Respir Crit Care Med. 2016 Aug;37(4):555-71. doi: 10.1055/s-0036-1584797. Epub 2016 Aug 3. Semin Respir Crit Care Med. 2016. PMID: 27486736 Free PMC article. Review.
-
Therapeutic antibodies, _targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice.iScience. 2021 May 21;24(5):102479. doi: 10.1016/j.isci.2021.102479. Epub 2021 Apr 26. iScience. 2021. PMID: 33937725 Free PMC article.
-
Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell.bioRxiv [Preprint]. 2021 Mar 1:2021.03.01.433431. doi: 10.1101/2021.03.01.433431. bioRxiv. 2021. Update in: Cell Rep. 2021 Jul 13;36(2):109364. doi: 10.1016/j.celrep.2021.109364 PMID: 33688646 Free PMC article. Updated. Preprint.
-
Author Correction: Multivalent 9-O-Acetylated-sialic acid glycoclusters as potent inhibitors for SARS-CoV-2 infection.Nat Commun. 2022 Jun 24;13(1):3611. doi: 10.1038/s41467-022-31290-8. Nat Commun. 2022. PMID: 35750667 Free PMC article. No abstract available.
-
A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.Natl Sci Rev. 2021 Mar 27;8(8):nwab053. doi: 10.1093/nsr/nwab053. eCollection 2021 Aug. Natl Sci Rev. 2021. PMID: 34676098 Free PMC article.
References
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen M-h, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus ADME, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. doi:10.1126/science.1085952. - DOI - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ; SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi:10.1056/NEJMoa030781. - DOI - PubMed
-
- Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA. 2013. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med 369:407–416. doi:10.1056/NEJMoa1306742. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

