Differential Regulation of NOTCH2 and NOTCH3 Contribute to Their Unique Functions in Vascular Smooth Muscle Cells
- PMID: 25957400
- PMCID: PMC4481222
- DOI: 10.1074/jbc.M115.655548
Differential Regulation of NOTCH2 and NOTCH3 Contribute to Their Unique Functions in Vascular Smooth Muscle Cells
Abstract
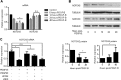
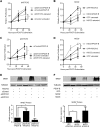
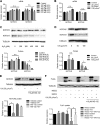
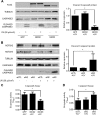
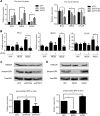
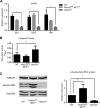
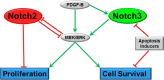
Notch signaling is a key regulator of vascular smooth muscle cell (VSMC) phenotypes, including differentiation, proliferation, and cell survival. However, the exact contribution of the individual Notch receptors has not been thoroughly delineated. In this study, we identify unique roles for NOTCH2 and NOTCH3 in regulating proliferation and cell survival in cultured VSMCs. Our results indicate that NOTCH2 inhibits PDGF-B-dependent proliferation and its expression is decreased by PDGF-B. In contrast, NOTCH3 promotes proliferation and receptor expression is increased by PDGF-B. Additionally, data show that NOTCH3, but not NOTCH2 protects VSMCs from apoptosis and apoptosis mediators degrade NOTCH3 protein. We identified three pro-survival genes specifically regulated by NOTCH3 in cultured VSMCs and in mouse aortas. This regulation is mediated through MAP kinase signaling, which we demonstrate can be activated by NOTCH3, but not NOTCH2. Overall, this study highlights discrete roles for NOTCH2 and NOTCH3 in VSMCs and connects these roles to specific upstream regulators that control their expression.
Keywords: Notch receptor; apoptosis; differentiation; mitogen-activated protein kinase (MAPK); proliferation; vascular smooth muscle cells.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Loss of Notch2 and Notch3 in vascular smooth muscle causes patent ductus arteriosus.Genesis. 2015 Dec;53(12):738-48. doi: 10.1002/dvg.22904. Epub 2015 Oct 30. Genesis. 2015. PMID: 26453897
-
A receptor-specific function for Notch2 in mediating vascular smooth muscle cell growth arrest through cyclin-dependent kinase inhibitor 1B.Circ Res. 2013 Sep 27;113(8):975-85. doi: 10.1161/CIRCRESAHA.113.301272. Epub 2013 Aug 21. Circ Res. 2013. PMID: 23965337 Free PMC article.
-
Notch2 and Notch3 function together to regulate vascular smooth muscle development.PLoS One. 2012;7(5):e37365. doi: 10.1371/journal.pone.0037365. Epub 2012 May 17. PLoS One. 2012. PMID: 22615991 Free PMC article.
-
An overview of Notch3 function in vascular smooth muscle cells.Prog Biophys Mol Biol. 2008 Jan-Apr;96(1-3):499-509. doi: 10.1016/j.pbiomolbio.2007.07.006. Epub 2007 Jul 29. Prog Biophys Mol Biol. 2008. PMID: 17854869 Review.
-
Notch3 signalling and vascular remodelling in pulmonary arterial hypertension.Clin Sci (Lond). 2019 Dec 20;133(24):2481-2498. doi: 10.1042/CS20190835. Clin Sci (Lond). 2019. PMID: 31868216 Free PMC article. Review.
Cited by
-
SM22α+ vascular mural cells are essential for vessel stability in tumors and undergo phenotype transition regulated by Notch signaling.J Exp Clin Cancer Res. 2020 Jul 2;39(1):124. doi: 10.1186/s13046-020-01630-x. J Exp Clin Cancer Res. 2020. PMID: 32616053 Free PMC article.
-
CircSOD2: A Novel Regulator for Smooth Muscle Proliferation and Neointima Formation.Arterioscler Thromb Vasc Biol. 2021 Dec;41(12):2961-2973. doi: 10.1161/ATVBAHA.121.316911. Epub 2021 Oct 21. Arterioscler Thromb Vasc Biol. 2021. PMID: 34670409 Free PMC article.
-
Mechanisms of Smooth Muscle Cell Differentiation Are Distinctly Altered in Thoracic Aortic Aneurysms Associated with Bicuspid or Tricuspid Aortic Valves.Front Physiol. 2017 Jul 25;8:536. doi: 10.3389/fphys.2017.00536. eCollection 2017. Front Physiol. 2017. PMID: 28790933 Free PMC article.
-
Association of NOTCH3 Gene Polymorphisms with Ischemic Stroke and its Subtypes: A Meta-Analysis.Medicina (Kaunas). 2019 Jul 8;55(7):351. doi: 10.3390/medicina55070351. Medicina (Kaunas). 2019. PMID: 31288479 Free PMC article.
-
Pericytes and vascular smooth muscle cells in central nervous system arteriovenous malformations.Front Physiol. 2023 Aug 4;14:1210563. doi: 10.3389/fphys.2023.1210563. eCollection 2023. Front Physiol. 2023. PMID: 37601628 Free PMC article. Review.
References
-
- Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 - PubMed
-
- Egan S. E., St-Pierre B., Leow C. C. (1998) Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr. Topics Microbiol. Immunol. 228, 273–324 - PubMed
-
- Bianchi S., Dotti M. T., Federico A. (2006) Physiology and pathology of notch signalling system. J. Cell. Physiol. 207, 300–308 - PubMed
-
- Pursglove S. E., Mackay J. P. (2005) CSL: a notch above the rest. Int. J. Biochem. Cell Biol. 37, 2472–2477 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

