Neural differentiation modulates the vertebrate brain specific splicing program
- PMID: 25993117
- PMCID: PMC4438066
- DOI: 10.1371/journal.pone.0125998
Neural differentiation modulates the vertebrate brain specific splicing program
Abstract
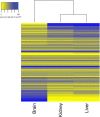
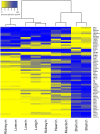
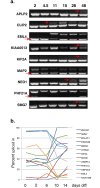
Alternative splicing patterns are known to vary between tissues but these patterns have been found to be predominantly peculiar to one species or another, implying only a limited function in fundamental neural biology. Here we used high-throughput RT-PCR to monitor the expression pattern of all the annotated simple alternative splicing events (ASEs) in the Reference Sequence Database, in different mouse tissues and identified 93 brain-specific events that shift from one isoform to another (switch-like) between brain and other tissues. Consistent with an important function, regulation of a core set of 9 conserved switch-like ASEs is highly conserved, as they have the same pattern of tissue-specific splicing in all vertebrates tested: human, mouse and zebrafish. Several of these ASEs are embedded within genes that encode proteins associated with the neuronal microtubule network, and show a dramatic and concerted shift within a short time window of human neural stem cell differentiation. Similarly these exons are dynamically regulated in zebrafish development. These data demonstrate that although alternative splicing patterns often vary between species, there is nonetheless a core set of vertebrate brain-specific ASEs that are conserved between species and associated with neural differentiation.
Conflict of interest statement
Figures


Similar articles
-
Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein.Cell. 2009 Sep 4;138(5):898-910. doi: 10.1016/j.cell.2009.06.012. Cell. 2009. PMID: 19737518
-
A novel protein domain in an ancestral splicing factor drove the evolution of neural microexons.Nat Ecol Evol. 2019 Apr;3(4):691-701. doi: 10.1038/s41559-019-0813-6. Epub 2019 Mar 4. Nat Ecol Evol. 2019. PMID: 30833759
-
HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens.Mol Vis. 2010 Nov 4;16:2301-16. Mol Vis. 2010. PMID: 21139978 Free PMC article.
-
Induction and alternative splicing of delta isoform of Ca(2+)/calmodulin-dependent protein kinase II during neural differentiation of P19 embryonal carcinoma cells and during brain development.Brain Res Mol Brain Res. 2000 Dec 28;85(1-2):189-99. doi: 10.1016/s0169-328x(00)00221-7. Brain Res Mol Brain Res. 2000. PMID: 11146121
-
Regulation of gene expression during early neuronal differentiation: evidence for patterns conserved across neuron populations and vertebrate classes.Cell Tissue Res. 2012 Apr;348(1):1-27. doi: 10.1007/s00441-012-1367-y. Epub 2012 Mar 23. Cell Tissue Res. 2012. PMID: 22437873 Review.
Cited by
-
Telomerase Activity is Downregulated Early During Human Brain Development.Genes (Basel). 2016 Jun 16;7(6):27. doi: 10.3390/genes7060027. Genes (Basel). 2016. PMID: 27322326 Free PMC article.
-
Zebrafish: an emerging real-time model system to study Alzheimer's disease and neurospecific drug discovery.Cell Death Discov. 2018 Oct 3;4:45. doi: 10.1038/s41420-018-0109-7. eCollection 2018. Cell Death Discov. 2018. PMID: 30302279 Free PMC article.
-
Hepatocellular senescence induces multi-organ senescence and dysfunction via TGFβ.Nat Cell Biol. 2024 Dec;26(12):2075-2083. doi: 10.1038/s41556-024-01543-3. Epub 2024 Nov 13. Nat Cell Biol. 2024. PMID: 39537753 Free PMC article.
-
Nutrition support use and clinical outcomes in patients with multiple myeloma undergoing autologous stem cell transplant.Support Care Cancer. 2022 Nov;30(11):9341-9350. doi: 10.1007/s00520-022-07358-y. Epub 2022 Sep 12. Support Care Cancer. 2022. PMID: 36089605
-
Modelling C9orf72-Related Amyotrophic Lateral Sclerosis in Zebrafish.Biomedicines. 2020 Oct 21;8(10):440. doi: 10.3390/biomedicines8100440. Biomedicines. 2020. PMID: 33096681 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

