Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy
- PMID: 26018823
- PMCID: PMC4590677
- DOI: 10.1080/15548627.2015.1017180
Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy
Abstract
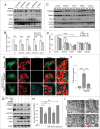
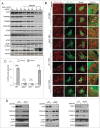
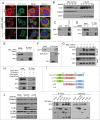
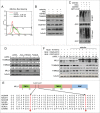
Mitochondria serve as membrane sources and signaling platforms for regulating autophagy. Accumulating evidence has also shown that damaged mitochondria are removed through both selective mitophagy and general autophagy in response to mitochondrial and oxidative stresses. Protein ubiquitination through mitochondrial E3 ligases plays an integrative role in mitochondrial outer membrane protein degradation, mitochondrial dynamics, and mitophagy. Here we showed that MUL1, a mitochondria-localized E3 ligase, regulates selenite-induced mitophagy in an ATG5 and ULK1-dependent manner. ULK1 partially translocated to mitochondria after selenite treatment and interacted with MUL1. We also demonstrated that ULK1 is a novel substrate of MUL1. These results suggest the association of mitochondria with autophagy regulation and provide a new mechanism for the beneficial effects of selenium as a chemopreventive agent.
Keywords: E3 ligase; MUL1; ULK1; mitophagy; selenite.
Figures


Similar articles
-
Thyroid hormone induction of mitochondrial activity is coupled to mitophagy via ROS-AMPK-ULK1 signaling.Autophagy. 2015;11(8):1341-57. doi: 10.1080/15548627.2015.1061849. Autophagy. 2015. PMID: 26103054 Free PMC article.
-
ULK1 and JNK are involved in mitophagy incurred by LRRK2 G2019S expression.Protein Cell. 2013 Sep;4(9):711-21. doi: 10.1007/s13238-013-3910-3. Epub 2013 Sep 10. Protein Cell. 2013. PMID: 27023913 Free PMC article.
-
The pro-oxidant adaptor p66SHC promotes B cell mitophagy by disrupting mitochondrial integrity and recruiting LC3-II.Autophagy. 2018;14(12):2117-2138. doi: 10.1080/15548627.2018.1505153. Epub 2018 Sep 6. Autophagy. 2018. PMID: 30109811 Free PMC article.
-
Mitochondrial E3 ubiquitin ligase 1: A key enzyme in regulation of mitochondrial dynamics and functions.Mitochondrion. 2016 May;28:49-53. doi: 10.1016/j.mito.2016.03.007. Epub 2016 Mar 24. Mitochondrion. 2016. PMID: 27034206 Review.
-
Autophagy machinery in the context of mammalian mitophagy.Biochim Biophys Acta. 2015 Oct;1853(10 Pt B):2797-801. doi: 10.1016/j.bbamcr.2015.01.013. Epub 2015 Jan 26. Biochim Biophys Acta. 2015. PMID: 25634658 Review.
Cited by
-
Role of the Mitochondrial E3 Ubiquitin Ligases as Possible Therapeutic _targets in Cancer Therapy.Int J Mol Sci. 2023 Dec 6;24(24):17176. doi: 10.3390/ijms242417176. Int J Mol Sci. 2023. PMID: 38139010 Free PMC article. Review.
-
Functions of autophagy in chloroplast protein degradation and homeostasis.Front Plant Sci. 2022 Sep 29;13:993215. doi: 10.3389/fpls.2022.993215. eCollection 2022. Front Plant Sci. 2022. PMID: 36247630 Free PMC article. Review.
-
Arabidopsis SINAT Proteins Control Autophagy by Mediating Ubiquitylation and Degradation of ATG13.Plant Cell. 2020 Jan;32(1):263-284. doi: 10.1105/tpc.19.00413. Epub 2019 Nov 15. Plant Cell. 2020. PMID: 31732704 Free PMC article.
-
BNIP3L/NIX-mediated mitophagy: molecular mechanisms and implications for human disease.Cell Death Dis. 2021 Dec 20;13(1):14. doi: 10.1038/s41419-021-04469-y. Cell Death Dis. 2021. PMID: 34930907 Free PMC article. Review.
-
Molecular Mechanisms and Regulation of Mammalian Mitophagy.Cells. 2021 Dec 23;11(1):38. doi: 10.3390/cells11010038. Cells. 2021. PMID: 35011599 Free PMC article. Review.
References
-
- Wallace DC, Brown MD, Melov S, Graham B, Lott M. Mitochondrial biology, degenerative diseases and aging. BioFactors 1998; 7:187-90; PMID:9568243; http://dx.doi.org/10.1002/biof.5520070303 - DOI - PubMed
-
- Green DR, Reed JC. Mitochondria and apoptosis. Science 1998; 281:1309-12; PMID:9721092; http://dx.doi.org/10.1126/science.281.5381.1309 - DOI - PubMed
-
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008; 283:10892-903; PMID:18281291; http://dx.doi.org/10.1074/jbc.M800102200 - DOI - PMC - PubMed
-
- Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res 2014; 24:42-57; PMID:24343578; http://dx.doi.org/10.1038/cr.2013.166 - DOI - PMC - PubMed
-
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. . Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456-61; PMID:21205641; http://dx.doi.org/10.1126/science.1196371 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
