Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas
- PMID: 26061751
- PMCID: PMC4530011
- DOI: 10.1056/NEJMoa1402121
Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas
Abstract
Background: Diffuse low-grade and intermediate-grade gliomas (which together make up the lower-grade gliomas, World Health Organization grades II and III) have highly variable clinical behavior that is not adequately predicted on the basis of histologic class. Some are indolent; others quickly progress to glioblastoma. The uncertainty is compounded by interobserver variability in histologic diagnosis. Mutations in IDH, TP53, and ATRX and codeletion of chromosome arms 1p and 19q (1p/19q codeletion) have been implicated as clinically relevant markers of lower-grade gliomas.
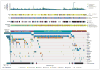
Methods: We performed genomewide analyses of 293 lower-grade gliomas from adults, incorporating exome sequence, DNA copy number, DNA methylation, messenger RNA expression, microRNA expression, and _targeted protein expression. These data were integrated and tested for correlation with clinical outcomes.
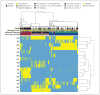
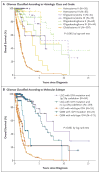
Results: Unsupervised clustering of mutations and data from RNA, DNA-copy-number, and DNA-methylation platforms uncovered concordant classification of three robust, nonoverlapping, prognostically significant subtypes of lower-grade glioma that were captured more accurately by IDH, 1p/19q, and TP53 status than by histologic class. Patients who had lower-grade gliomas with an IDH mutation and 1p/19q codeletion had the most favorable clinical outcomes. Their gliomas harbored mutations in CIC, FUBP1, NOTCH1, and the TERT promoter. Nearly all lower-grade gliomas with IDH mutations and no 1p/19q codeletion had mutations in TP53 (94%) and ATRX inactivation (86%). The large majority of lower-grade gliomas without an IDH mutation had genomic aberrations and clinical behavior strikingly similar to those found in primary glioblastoma.
Conclusions: The integration of genomewide data from multiple platforms delineated three molecular classes of lower-grade gliomas that were more concordant with IDH, 1p/19q, and TP53 status than with histologic class. Lower-grade gliomas with an IDH mutation either had 1p/19q codeletion or carried a TP53 mutation. Most lower-grade gliomas without an IDH mutation were molecularly and clinically similar to glioblastoma. (Funded by the National Institutes of Health.).
Figures



Comment in
-
Multiple Molecular Data Sets and the Classification of Adult Diffuse Gliomas.N Engl J Med. 2015 Jun 25;372(26):2555-7. doi: 10.1056/NEJMe1506813. Epub 2015 Jun 10. N Engl J Med. 2015. PMID: 26061754 No abstract available.
-
CNS cancer: molecular classification of glioma.Nat Rev Clin Oncol. 2015 Sep;12(9):502. doi: 10.1038/nrclinonc.2015.111. Epub 2015 Jul 14. Nat Rev Clin Oncol. 2015. PMID: 26169923 No abstract available.
Similar articles
-
TERT promoter mutation confers favorable prognosis regardless of 1p/19q status in adult diffuse gliomas with IDH1/2 mutations.Acta Neuropathol Commun. 2020 Nov 23;8(1):201. doi: 10.1186/s40478-020-01078-2. Acta Neuropathol Commun. 2020. PMID: 33228806 Free PMC article.
-
Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors.N Engl J Med. 2015 Jun 25;372(26):2499-508. doi: 10.1056/NEJMoa1407279. Epub 2015 Jun 10. N Engl J Med. 2015. PMID: 26061753 Free PMC article.
-
RNA editing-based classification of diffuse gliomas: predicting isocitrate dehydrogenase mutation and chromosome 1p/19q codeletion.BMC Bioinformatics. 2019 Dec 24;20(Suppl 19):659. doi: 10.1186/s12859-019-3236-0. BMC Bioinformatics. 2019. PMID: 31870275 Free PMC article.
-
Biomarker-driven diagnosis of diffuse gliomas.Mol Aspects Med. 2015 Nov;45:87-96. doi: 10.1016/j.mam.2015.05.002. Epub 2015 May 21. Mol Aspects Med. 2015. PMID: 26004297 Review.
-
Integrated diagnostics of diffuse astrocytic and oligodendroglial tumors.Pathologe. 2019 Jun;40(Suppl 1):9-17. doi: 10.1007/s00292-019-0581-8. Pathologe. 2019. PMID: 31025086 Review. English.
Cited by
-
Survival Determinants in Glioblastoma: An Insight into Biopsy-Only Patient Outcomes.Biomedicines. 2024 Oct 13;12(10):2327. doi: 10.3390/biomedicines12102327. Biomedicines. 2024. PMID: 39457639 Free PMC article.
-
Current Approaches for Glioma Gene Therapy and Virotherapy.Front Mol Neurosci. 2021 Mar 11;14:621831. doi: 10.3389/fnmol.2021.621831. eCollection 2021. Front Mol Neurosci. 2021. PMID: 33790740 Free PMC article. Review.
-
CENP-A is a potential prognostic biomarker and correlated with immune infiltration levels in glioma patients.Front Genet. 2022 Aug 29;13:931222. doi: 10.3389/fgene.2022.931222. eCollection 2022. Front Genet. 2022. PMID: 36105094 Free PMC article.
-
Sterol Regulatory Element Binding Protein Regulates the Expression and Metabolic Functions of Wild-Type and Oncogenic IDH1.Mol Cell Biol. 2016 Aug 26;36(18):2384-95. doi: 10.1128/MCB.00163-16. Print 2016 Sep 15. Mol Cell Biol. 2016. PMID: 27354064 Free PMC article.
-
Identification of Novel Transcriptome Signature as a Potential Prognostic Biomarker for Anti-Angiogenic Therapy in Glioblastoma Multiforme.Cancers (Basel). 2021 Mar 1;13(5):1013. doi: 10.3390/cancers13051013. Cancers (Basel). 2021. PMID: 33804433 Free PMC article.
References
-
- Macdonald DR, Gaspar LE, Cairn-cross JG. Successful chemotherapy for newly diagnosed aggressive oligodendroglioma. Ann Neurol. 1990;27:573–4. - PubMed
-
- van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24CA143840/CA/NCI NIH HHS/United States
- U24CA126561/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24CA143731/CA/NCI NIH HHS/United States
- U24CA126551/CA/NCI NIH HHS/United States
- U24CA126543/CA/NCI NIH HHS/United States
- U24CA126554/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- U24CA143858/CA/NCI NIH HHS/United States
- U24CA126544/CA/NCI NIH HHS/United States
- P30CA016672/CA/NCI NIH HHS/United States
- U24CA143882/CA/NCI NIH HHS/United States
- U24 CA126551/CA/NCI NIH HHS/United States
- U24CA144025/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- U01 CA168394/CA/NCI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- U24 CA194362/CA/NCI NIH HHS/United States
- U24CA143883/CA/NCI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- U24 CA126554/CA/NCI NIH HHS/United States
- U24 CA180951/CA/NCI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- U24CA143867/CA/NCI NIH HHS/United States
- U24CA143835/CA/NCI NIH HHS/United States
- U24 CA126561/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24CA143843/CA/NCI NIH HHS/United States
- R01 NS081125/NS/NINDS NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U24 CA126543/CA/NCI NIH HHS/United States
- U24CA143848/CA/NCI NIH HHS/United States
- U24CA126563/CA/NCI NIH HHS/United States
- U24CA126546/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24CA143799/CA/NCI NIH HHS/United States
- U54 HG007990/HG/NHGRI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- R01 CA163722/CA/NCI NIH HHS/United States
- U24CA143866/CA/NCI NIH HHS/United States
- U24CA143845/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA126546/CA/NCI NIH HHS/United States
- U24 CA126563/CA/NCI NIH HHS/United States
- U24 CA126544/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
