Manganese lipoxygenase of F. oxysporum and the structural basis for biosynthesis of distinct 11-hydroperoxy stereoisomers
- PMID: 26113537
- PMCID: PMC4514001
- DOI: 10.1194/jlr.M060178
Manganese lipoxygenase of F. oxysporum and the structural basis for biosynthesis of distinct 11-hydroperoxy stereoisomers
Abstract
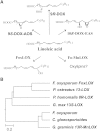
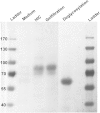
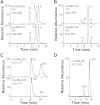
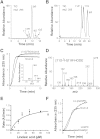
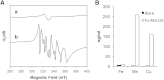
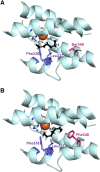
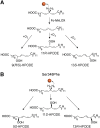
The biosynthesis of jasmonates in plants is initiated by 13S-lipoxygenase (LOX), but details of jasmonate biosynthesis by fungi, including Fusarium oxysporum, are unknown. The genome of F. oxysporum codes for linoleate 13S-LOX (FoxLOX) and for F. oxysporum manganese LOX (Fo-MnLOX), an uncharacterized homolog of 13R-MnLOX of Gaeumannomyces graminis. We expressed Fo-MnLOX and compared its properties to Cg-MnLOX from Colletotrichum gloeosporioides. Electron paramagnetic resonance and metal analysis showed that Fo-MnLOX contained catalytic Mn. Fo-MnLOX oxidized 18:2n-6 mainly to 11R-hydroperoxyoctadecadienoic acid (HPODE), 13S-HPODE, and 9(S/R)-HPODE, whereas Cg-MnLOX produced 9S-, 11S-, and 13R-HPODE with high stereoselectivity. The 11-hydroperoxides did not undergo the rapid β-fragmentation earlier observed with 13R-MnLOX. Oxidation of [11S-(2)H]18:2n-6 by Cg-MnLOX was accompanied by loss of deuterium and a large kinetic isotope effect (>30). The Fo-MnLOX-catalyzed oxidation occurred with retention of the (2)H-label. Fo-MnLOX also oxidized 1-lineoyl-2-hydroxy-glycero-3-phosphatidylcholine. The predicted active site of all MnLOXs contains Phe except for Ser(348) in this position of Fo-MnLOX. The Ser348Phe mutant of Fo-MnLOX oxidized 18:2n-6 to the same major products as Cg-MnLOX. Our results suggest that Fo-MnLOX, with support of Ser(348), binds 18:2n-6 so that the proR rather than the proS hydrogen at C-11 interacts with the metal center, but retains the suprafacial oxygenation mechanism observed in other MnLOXs.
Keywords: Fusarium gloeosporioides; Fusarium oxysporum; Pichia pastoris; gene expression; mass spectrometry; oxygenation mechanism; oxylipins; yeast expression.
Copyright © 2015 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Secretion of two novel enzymes, manganese 9S-lipoxygenase and epoxy alcohol synthase, by the rice pathogen Magnaporthe salvinii.J Lipid Res. 2013 Mar;54(3):762-775. doi: 10.1194/jlr.M033787. Epub 2012 Dec 11. J Lipid Res. 2013. PMID: 23233731 Free PMC article.
-
Iron and manganese lipoxygenases of plant pathogenic fungi and their role in biosynthesis of jasmonates.Arch Biochem Biophys. 2022 Jun 15;722:109169. doi: 10.1016/j.abb.2022.109169. Epub 2022 Mar 8. Arch Biochem Biophys. 2022. PMID: 35276213 Review.
-
Expression and characterization of manganese lipoxygenase of the rice blast fungus reveals prominent sequential lipoxygenation of α-linolenic acid.Arch Biochem Biophys. 2015 Oct 1;583:87-95. doi: 10.1016/j.abb.2015.07.014. Epub 2015 Aug 8. Arch Biochem Biophys. 2015. PMID: 26264916
-
Crystal structure of linoleate 13R-manganese lipoxygenase in complex with an adhesion protein.J Lipid Res. 2016 Aug;57(8):1574-88. doi: 10.1194/jlr.M069617. Epub 2016 Jun 15. J Lipid Res. 2016. PMID: 27313058 Free PMC article.
-
Plant and fungal lipoxygenases.Prostaglandins Other Lipid Mediat. 2002 Aug;68-69:313-23. doi: 10.1016/s0090-6980(02)00037-0. Prostaglandins Other Lipid Mediat. 2002. PMID: 12432926 Review.
Cited by
-
Crystal Structure of Manganese Lipoxygenase of the Rice Blast Fungus Magnaporthe oryzae.J Biol Chem. 2016 Apr 8;291(15):8130-9. doi: 10.1074/jbc.M115.707380. Epub 2016 Jan 18. J Biol Chem. 2016. PMID: 26783260 Free PMC article.
-
The green microalga Lobosphaera incisa harbours an arachidonate 15S-lipoxygenase.Plant Biol (Stuttg). 2019 Jan;21 Suppl 1(Suppl Suppl 1):131-142. doi: 10.1111/plb.12920. Epub 2018 Oct 24. Plant Biol (Stuttg). 2019. PMID: 30277010 Free PMC article.
-
Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.Sci Rep. 2017 May 18;7(1):2069. doi: 10.1038/s41598-017-02153-w. Sci Rep. 2017. PMID: 28522865 Free PMC article.
-
HAA by the first {Mn(iii)OH} complex with all O-donor ligands.Chem Sci. 2023 Jul 12;14(30):8187-8195. doi: 10.1039/d3sc01971c. eCollection 2023 Aug 2. Chem Sci. 2023. PMID: 37538819 Free PMC article.
-
Extracellular lipopeptide bacillomycin L regulates serial expression of genes for modulating multicellular behavior in Bacillus velezensis Bs916.Appl Microbiol Biotechnol. 2021 Sep;105(18):6853-6870. doi: 10.1007/s00253-021-11524-3. Epub 2021 Sep 3. Appl Microbiol Biotechnol. 2021. PMID: 34477941
References
-
- Minor W., Steczko J., Stec B., Otwinowski Z., Bolin J. T., Walter R., Axelrod B. 1996. Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution. Biochemistry. 35: 10687–10701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

