Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain
- PMID: 26141956
- PMCID: PMC4936782
- DOI: 10.1016/j.ydbio.2015.06.021
Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain
Abstract
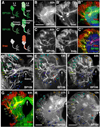
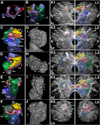
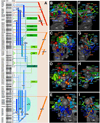
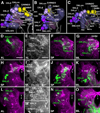
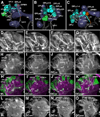
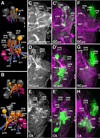
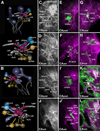
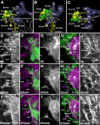
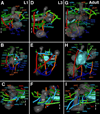
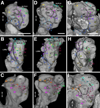
Fixed lineages derived from unique, genetically specified neuroblasts form the anatomical building blocks of the Drosophila brain. Neurons belonging to the same lineage project their axons in a common tract, which is labeled by neuronal markers. In this paper, we present a detailed atlas of the lineage-associated tracts forming the brain of the early Drosophila larva, based on the use of global markers (anti-Neuroglian, anti-Neurotactin, inscuteable-Gal4>UAS-chRFP-Tub) and lineage-specific reporters. We describe 68 discrete fiber bundles that contain axons of one lineage or pairs/small sets of adjacent lineages. Bundles enter the neuropil at invariant locations, the lineage tract entry portals. Within the neuropil, these fiber bundles form larger fascicles that can be classified, by their main orientation, into longitudinal, transverse, and vertical (ascending/descending) fascicles. We present 3D digital models of lineage tract entry portals and neuropil fascicles, set into relationship to commonly used, easily recognizable reference structures such as the mushroom body, the antennal lobe, the optic lobe, and the Fasciclin II-positive fiber bundles that connect the brain and ventral nerve cord. Correspondences and differences between early larval tract anatomy and the previously described late larval and adult lineage patterns are highlighted. Our L1 neuro-anatomical atlas of lineages constitutes an essential step towards following morphologically defined lineages to the neuroblasts of the early embryo, which will ultimately make it possible to link the structure and connectivity of a lineage to the expression of genes in the particular neuroblast that gives rise to that lineage. Furthermore, the L1 atlas will be important for a host of ongoing work that attempts to reconstruct neuronal connectivity at the level of resolution of single neurons and their synapses.
Keywords: Brain; Development; Drosophila; Larval; Lineage.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Structure and development of the subesophageal zone of the Drosophila brain. I. Segmental architecture, compartmentalization, and lineage anatomy.J Comp Neurol. 2018 Jan 1;526(1):6-32. doi: 10.1002/cne.24287. Epub 2017 Aug 10. J Comp Neurol. 2018. PMID: 28730682 Free PMC article.
-
Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage.J Neurosci. 2006 May 17;26(20):5534-53. doi: 10.1523/JNEUROSCI.4708-05.2006. J Neurosci. 2006. PMID: 16707805 Free PMC article.
-
Early development of the Drosophila brain: V. Pattern of postembryonic neuronal lineages expressing DE-cadherin.J Comp Neurol. 2003 Jan 20;455(4):451-62. doi: 10.1002/cne.10484. J Comp Neurol. 2003. PMID: 12508319
-
Embryonic development of the insect central complex: insights from lineages in the grasshopper and Drosophila.Arthropod Struct Dev. 2011 Jul;40(4):334-48. doi: 10.1016/j.asd.2011.02.005. Epub 2011 Mar 5. Arthropod Struct Dev. 2011. PMID: 21382507 Review.
-
Wiring the Drosophila Brain with Individually Tailored Neural Lineages.Curr Biol. 2017 Jan 23;27(2):R77-R82. doi: 10.1016/j.cub.2016.12.026. Curr Biol. 2017. PMID: 28118595 Review.
Cited by
-
A genome-wide resource for the analysis of protein localisation in Drosophila.Elife. 2016 Feb 20;5:e12068. doi: 10.7554/eLife.12068. Elife. 2016. PMID: 26896675 Free PMC article.
-
Structure and development of the subesophageal zone of the Drosophila brain. II. Sensory compartments.J Comp Neurol. 2018 Jan 1;526(1):33-58. doi: 10.1002/cne.24316. Epub 2017 Sep 28. J Comp Neurol. 2018. PMID: 28875566 Free PMC article.
-
Development of the anterior visual input pathway to the Drosophila central complex.J Comp Neurol. 2017 Nov 1;525(16):3458-3475. doi: 10.1002/cne.24277. Epub 2017 Aug 21. J Comp Neurol. 2017. PMID: 28675433 Free PMC article.
-
Structure and development of the subesophageal zone of the Drosophila brain. I. Segmental architecture, compartmentalization, and lineage anatomy.J Comp Neurol. 2018 Jan 1;526(1):6-32. doi: 10.1002/cne.24287. Epub 2017 Aug 10. J Comp Neurol. 2018. PMID: 28730682 Free PMC article.
-
Developmentally Arrested Precursors of Pontine Neurons Establish an Embryonic Blueprint of the Drosophila Central Complex.Curr Biol. 2019 Feb 4;29(3):412-425.e3. doi: 10.1016/j.cub.2018.12.012. Epub 2019 Jan 17. Curr Biol. 2019. PMID: 30661802 Free PMC article.
References
-
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. - PubMed
-
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

